On April 16, BGI-Research announced the publication of two joint studies conducted by the Chinese Academy of Sciences Center for Excellence in Molecular Cell Science (CEMCS) in collaboration with the Jilin University, IRCCS Istituto Tumori "Giovanni Paolo II" of Bari (Italy), University of Copenhagen, Chulalongkorn University, University of Cambridge, Hannover Medical School, and other Chinese and international institutions. Both studies employed BGI's proprietary high-resolution and large field-of-view spatial multi-omics technology (Stereo-seq) in combination with single-cell transcriptome sequencing technology, resulting in ground-breaking discoveries that were published online today and will be in print on the May issue of Nature Genetics.
The main achievement was the generation of a comprehensive high-definition spatiotemporal atlas of the mouse liver, which offers a detailed, spatially-resolved, understanding of the liver’s molecular features under normal conditions and how those are changing in regenerative conditions following injury. From the wider perspective, these findings provide new insights in molecular pathways that can be exploited for improving the treatment of various liver diseases as well as promoting liver regeneration and improving transplantation procedures.


Both studies “A spatiotemporal atlas of mouse liver homeostasis and regeneration” and “A spatiotemporal atlas of cholestatic injury and repair in mice” were published in Nature Genetics.
A spatiotemporal atlas of mouse liver homeostasis and regeneration
The liver serves multiple essential functions in the body, not only detoxifying harmful substances but also carrying out metabolic, synthetic, digestive, and immune functions. To accomplish those tasks, different types of cells in various spatial regions of the liver work together in a coordinated manner. The spatial distribution of liver functions is of great significance in understanding the development of liver diseases and the process of liver regeneration after injury. The specific location of cells within the liver is closely tied to the heterogeneity observed in the progression of liver diseases and the mechanisms underlying liver regeneration.
In modern lifestyles, characterized by long hours of late-night work, smoking, excessive alcohol consumption, as well as high-sugar and high-salt diets, liver diseases are becoming a growing concern at an alarming rate. To address this challenge, it is paramount to resolve the mechanisms that govern liver cell damage and regeneration in the spatial dimension. Such understanding is essential for determining how liver diseases transform into malignant forms and in advancing the field of regenerative medicine.
The liver lobules are the fundamental units that make up the structure and function of the mammalian liver. An adult liver typically contains between 500,000 to 1,000,000 liver lobules. Within each lobule, there is a flow of blood from the portal area to the central vein area. This creates a gradient of various substances such as nutrients, hormones, cells, and growth factors. This phenomenon is known as liver zonation, which refers to the distinct biological functions exhibited by cells at different spatial locations within the lobule. By understanding liver zonation, we gain insights into how different regions within the liver contribute to its overall functionality.
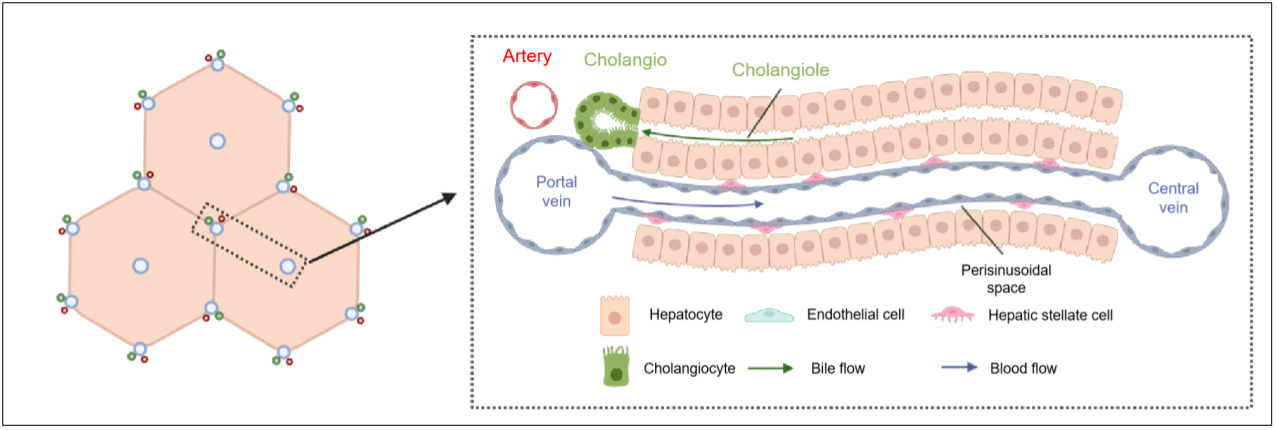 The structure of liver lobule. (By Yue Jin, CEMCS)
The structure of liver lobule. (By Yue Jin, CEMCS)
Led by BGI-Research and in collaboration with Jilin University, CEMCS, the Fifth Affiliated Hospital of Guangzhou Medical University, Shanxi Medical University, IRCCS-Istituto Tumori, and Hannover Medical School, the study “A spatiotemporal atlas of mouse liver homeostasis and regeneration” explored the mechanisms of liver homeostasis and regeneration after partial liver resection.
Employing a cutting-edge technique called Stereo-seq, the research team conducted their study on both healthy homeostatic livers and livers that underwent 70% resection in mice, constructing a detailed spatiotemporal network of liver lobules with an incredible resolution at the nanometer scale. Such approached allowed the research team to uncover the spatial characteristics of cell types, gene expression patterns, and microenvironmental signals within the steady-state mouse liver, as well as the coordinated spatiotemporal reshaping of the transcriptome following partial liver resection. These dynamic processes play a vital role in accurately controlling liver and orchestrating the regeneration of the liver.
The research team unveiled a previously unknown immune microstructure residing within the liver, ranging from 50 to 200 micrometers and primarily consisting of T cells (a type of white blood cell) and monocytes (a type of leukocyte or white blood cell). This newly identified immune microstructure is believed to play a significant role in the liver's immune response. It is speculated that this unique mechanism serves as a defense mechanism, aiding the liver in combating external bacteria or antigens that may pose a threat to its proper functioning.
The team conducted a detailed analysis of gene expression patterns in various types of liver cells, taking into account their specific locations within the liver zonation. They identified a set of genes that exhibit regional distribution and are closely related to diseases such as hepatitis B.
By analyzing the liver regenerative capacity partial resection, a number of key transcription factors were identified and validated, including those involved in maintaining liver zonation patterns. Moreover, they also elucidated the coordinated spatiotemporal mechanisms that govern metabolic activities and gene regulatory networks that orchestrate the complex process of liver regeneration.
Dr. Yiwei Lai, one of the co-corresponding authors from BGI-Research, commented, "This research highlights BGI's strengths in advanced technologies such as Stereo-seq, and provides a valuable data resource for understanding liver homeostasis and regeneration. Furthermore, it establishes a critical foundation for further investigations into the physiology and malfunction of the mammalian liver."
A spatiotemporal atlas of cholestatic injury and repair in mice
Cholestasis is a condition characterized by a disruption in the normal bile flow through the biliary system, which can result in liver damage. This condition hampers bile secretion and excretion, leading to serious diseases such as cholecystitis, liver cirrhosis, and liver failure.
Previous studies revealed that after cholestatic liver injury, specialized cells around the portal vein transform into liver progenitor-like cells (LPLCs). These cells possess the ability to develop into mature hepatocytes (liver cells) and cholangiocytes (epithelial cells in the bile ducts) with high proliferative ability, those contributing to liver regeneration.
During the initial phase of liver repair, extensive hepatocyte proliferation occurs primarily in the region surrounding the central vein. However, the precise mechanisms that control the proliferation process are not yet fully understood.
In the study “A spatiotemporal atlas of cholestatic injury and repair in mice," CEMCS, in partnership with BGI-Research and other institutes, have created a comprehensive spatiotemporal transcriptomic atlas of liver injury and regeneration caused by cholestasis. This atlas reveals critical information about the spatiotemporal dynamics of cellular responses to injury and microenvironmental signals involved.
To simulate human cholestatic injury, the research team induced region-specific damage around the portal veins, while leaving the areas near the central veins undamaged. By analyzing the interactions among the cells in the portal vein area, the team discovered that bile duct cells play a crucial role as signaling hubs under cholestasis. They integrate cell interactions within the microenvironment of the portal vein area and are closely related to the reshaping of regional immune cells and hepatocyte reprogramming.
Utilizing BGI's Stereo-seq technology, the team identified two distinct subtypes of LPLCs during the hepatocyte reprogramming process, with one subtype exhibiting a stronger tendency towards bile duct differentiation closer to the bile duct area. The study also identified key factors that can limit hepatocyte proliferation, providing new insights and potential targets for understanding region-specific liver injury and the treatment of bile duct-related diseases.
"The findings from this study not only provide valuable data resources for understanding regional liver injury but also potential intervention strategies for promoting hepatocyte regeneration under cholestasis," stated Dr. Lijian Hui, co-corresponding author and a principal investigator at the CEMCS.
Together, both studies form a comprehensive liver reference atlas that addresses various aspects of liver injury and regeneration. These findings have the potential to uncover underlying mechanisms and targets for promoting the regeneration of region-specific liver injuries. Moreover, they establish a new research framework for studying the intricate mechanisms of injury and regeneration in other tissues that exhibit heterogeneous distribution patterns.
The studies can be accessed here:
A spatiotemporal atlas of mouse liver homeostasis and regeneration
https://www.nature.com/articles/s41588-024-01709-7
A spatiotemporal atlas of cholestatic injury and repair in mice
https://www.nature.com/articles/s41588-024-01687-w



