Bats and rodents play a critical role in the transmission of zoonotic pathogens, closely associated with the spread of Ebola, Marburg virus, and SARS-CoV, which pose significant health and economic threats globally. Countries and regions facing the biggest challenges are those with weak public health facilities, a lack of medical resources, and poor environmental conditions, disease surveillance and control. Investigating the evolution patterns and transmission dynamics of viruses in wild animal populations in these areas constitutes a pivotal stride in preemptively identifying and mitigating future pandemic risks.
To help address these challenges, BGI-Research, in collaboration with the Wuhan Institute of Virology at the Chinese Academy of Sciences, the University of Queensland, and other institutes, conducted a large-scale virome survey of 959 bats and 372 rodents in Kenya and Uganda. The research team uncovered the viral diversity, recombination, and cross-regional transmission characteristics in these wildlife species through metatranscriptomic sequencing. The research findings are published in the international scientific journal Microbiome on April 10th.
 The research “Substantial viral diversity in bats and rodents from East Africa: insights into evolution, recombination, and cocirculation” is published in Microbiome.
The research “Substantial viral diversity in bats and rodents from East Africa: insights into evolution, recombination, and cocirculation” is published in Microbiome.
The research team identified a total of 251 RNA and DNA viruses related to bat or rodent infections, of which 87% are new viruses, revealing the broad diversity and uniqueness in the East African region. Notably, viruses closely related to human parainfluenza virus 2 (HPIV-2) and human molluscum contagiosum virus (MCV) were found in Egyptian fruit bats. These findings are not only significant for understanding the origins of these viruses but also provide crucial insights into their transmission mechanisms.
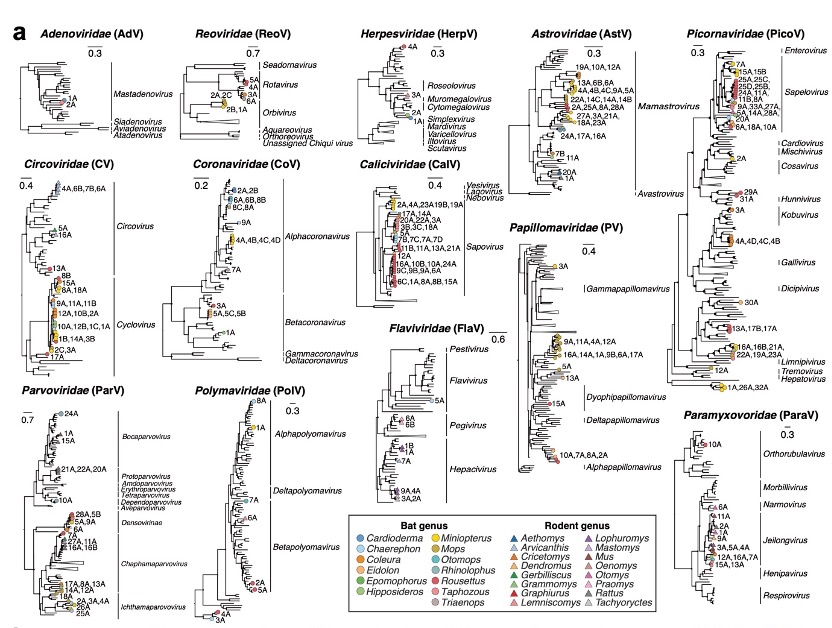 Viral diversity in East African bats and rodents.
Viral diversity in East African bats and rodents.
Furthermore, the study, through comparative analysis among viral families, revealed that Coronaviruses and Circoviruses exhibit high diversity on spike and capsid proteins (responsible for invading host cells), and frequent recombination event occurs between viruses. In short, these viruses attach to cells, enter, replicate, and then leave to infect more cells. This diversity and flexible combination allow them to attack a wider variety of hosts, thereby having a broader range of transmission.
Further analysis has revealed the diversity of viral communities, which is a result of several key factors working together. First, frequent recurrent mutations within the viral communities increase their genetic diversity. Second, the co-infection of different viral types indicates that although viruses may belong to the same species, there are different types within this virus. Additionally, the viral flow between regions further promotes the diversification of viral communities. These factors together enhance the diversity of viral communities in nature, providing important information for us to understand how viruses spread in different ecosystems.
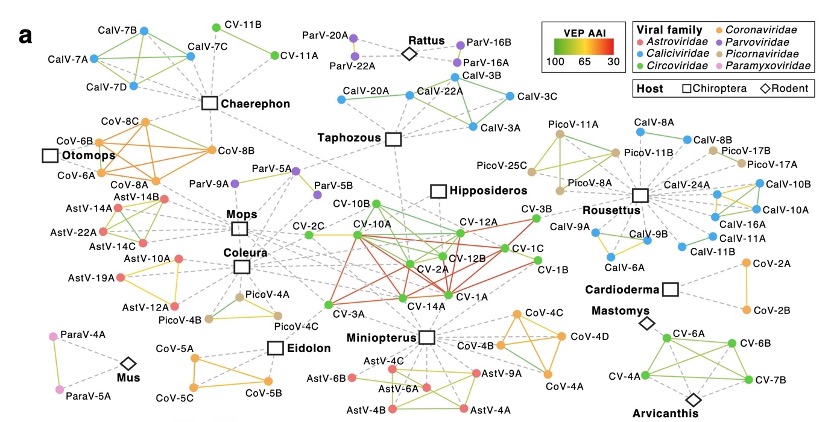 Network of host range and genome variability.
Network of host range and genome variability.
This research not only provides new genomic data and perspectives for global viral diversity research, but also makes an important contribution to global public health security, which closely associates with the "One Health" concept.
This study is based on the Global Pathogen Database Project (GPDP), which aims to detect and discover pathogens in animals that could potentially infect humans at the genomic level. By the end of 2026, GPDP aims to support research teams in collaborating to obtain pathogen genomic data from at least 50,000 animal-derived samples, in order to identify new zoonotic pathogens, identify hotspots of pathogen distribution, and predict potential cross-species transmission events.
Dr. Daxi WANG, Director of the Global Pathogen Database Project from BGI-Research, stated, "This research provides comprehensive genomic and ecological data resources for the virus spectrum of important pathogen reservoirs. Our team will continue to develop pathogen data platform, integrating multidimensional information including various pathogens, wildlife, and ecology. Those efforts will provide solid data support for infectious disease prevention, as well as surveillance and early warning of emerging diseases.”
Ethical approval was obtained for this research.
The research can be accessed here:
https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-024-01782-4
Source:
UN World Health Organization: One Health
https://www.who.int/news-room/questions-and-answers/item/one-health



