The effort of teams from six countries – including BGI representing China - over 13 years to complete the Human Genome Project revolutionized the development of life sciences and medicine. However, the completion of the Human Genome Project was really just the beginning for the next phase of scientific research. Recent breakthroughs in single-cell omics and spatiotemporal omics have provided new insights into the genome and brought new opportunities for biological and medical research.
To mark the 50th anniversary of the scientific journal Cell, Dr. Xu Xun, Director of BGI-Research, along with a team from BGI-Research, was invited to contribute a scientific review. They focused on the technical development and challenges of spatiotemporal omics and its potential to enhance our understanding of the human genome. This review was published on August 22 as a special issue in Cell.
 The Review "Spatiotemporal omics for biology and medicine" was published in Cell.
The Review "Spatiotemporal omics for biology and medicine" was published in Cell.
A Study of Life
The completion of the Human Genome Project unraveled 3 billion DNA base pairs and approximately 25,000 genes in the human genetic code, opening an era of studying how genetic information determines biological function, helping to uncover the differences between humans and other species, and the basis of genetic variation behind a large number of phenotypes and diseases.
The first challenge in understanding the genome is how the 3 billion base pairstranslate into a complete life. For example, how does a fertilized egg, which contains complete genomic information, eventually develop into a complex individual and age further over time? The second challenge is, how does the genetic variant associated with phenotype and inherited disease relate to our tissues and organs? Genomic analyses based on large populations, such as the 1000 Genomes Project and genome-wide association studies (GWAS), have revealed numerous variants associated with human phenotypes and diseases, but much remains unknown about how these variants specifically affect the function of specific organs or cells.
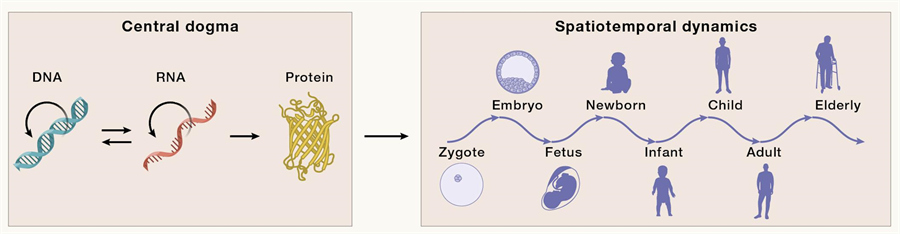 From Central dogma to the study of Spatiotemporal dynamics of life
From Central dogma to the study of Spatiotemporal dynamics of life
The adult human body contains about 37 trillion cells that are arranged in a specific way to form organs and tissues for various functions. A comprehensive understanding of the function of these organs or tissues requires a multidimensional analysis of the cells, including:
· Qualitative - cell type diversity resulting from the regulation of gene expression;
· Quantitative - count, ratio, and density of each cell type;
· Space - the spatial arrangement of each cell type and its interactions;
· Time - the time point of cell type and state change;
· Direction - the potential transformation trajectories that each cell type may undergo.
Taken together, these dimensions will eventually form a standard for the definition of the cell. Understanding these dimensions of cellular diversity and organization is critical to unraveling the connections between the genome and specific biological processes and their regulatory mechanisms.
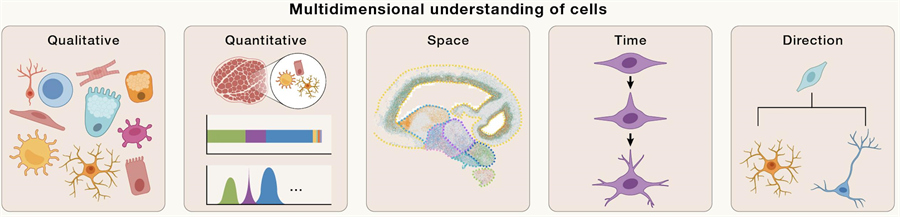 Multidimensional dissection and definition of cells
Multidimensional dissection and definition of cells
From genomics, cell omics to spatial omics
Advances in sequencing technology, especially the advent of Sequencing-By -Synthesis (SBS) technology, have made large-scale multi-omics analysis possible. These methods can analyze the regulatory heterogeneity of genetic information expression at the tissue level, but they cannot fully reveal the heterogeneity of different cell types within tissues.
The rapid development of single-cell sequencing technology has provided unprecedented tools for analyzing cellular heterogeneity within tissues. Single-cell omics can define cell types and states in multiple dimensions, including genome, transcriptome, epigenome, and proteome, and has become a key technical means to decode how genomic information is transcribed and converted into information about specific cell types.
Single-cell omics, however, lacks location information. Meanwhile, traditional imaging methods such as X-ray, computed tomography (CT), and magnetic resonance imaging (MRI) can visualize the 3D structure of tissues and organs, but lack molecular and cellular resolution. Immunohistochemistry (IHC) or in situ hybridization (ISH) allows for the spatial localization of a particular gene or protein, but only a limited number of targets can be detected.
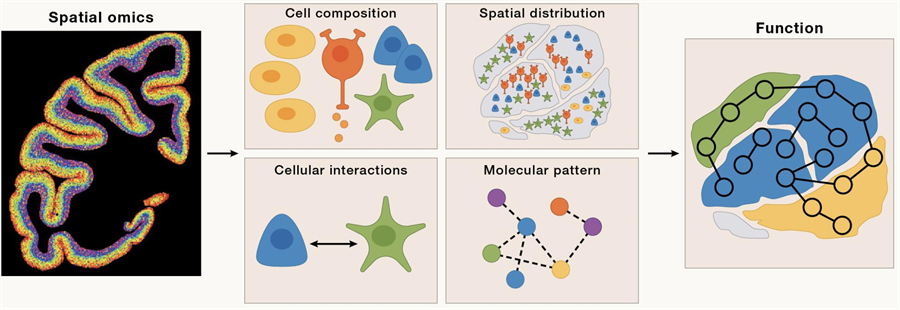
It is only through the development of spatial omics technology that scientists have been able to comprehensively map cell composition, localization, cell-cell interactions, and spatial dynamics of cellular ecosystems. From a functional perspective, these variables are critical for understanding morphogenesis during development, the structure of different organs and their subsequent functional changes, and the changes in the cellular microenvironment associated with disease processes.
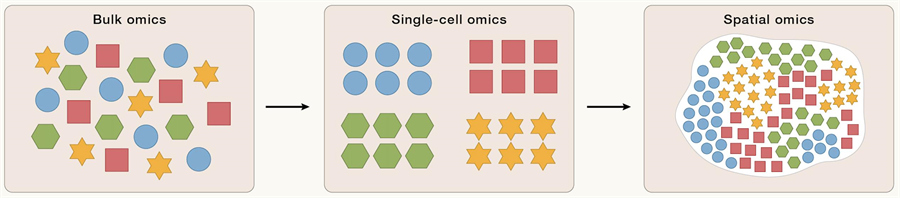 Genomics (DNA),cell omics, and spatial omics (DCS)
Genomics (DNA),cell omics, and spatial omics (DCS)
Techniques and Algorithms for Spatiotemporal Omics
Fundamentally, spatial omics is the simultaneous detection of a large number of molecules and their spatial positions. In situ imaging-based spatial omics techniques, such as fluorescence in situ hybridization (FISH) and in situ sequencing (ISS), enable scientists to directly observe the distribution of RNA and DNA within cells. Earlier techniques, such as smFISH, have enabled high-resolution imaging of individual RNA molecules. With the development of technology, modern spatial omics techniques such as MERFISH and seqFISH can detect the expression of thousands of genes simultaneously in a single cell.
Sequencing-based spatial omics technology enables genome-wide analysis. With the advent of high-throughput labeling methods, spatial omics technology has further developed from the early micron-level resolution technologies such as Visium, DBiT-seq, and Slide-seq to nanometer-level resolution technologies such as Stereo-seq, Seq-scope, and Pixel-seq.
Another important aspect of spatial omics technology is its integration with other omics techniques, such as proteomics and metabolomics, to provide a more comprehensive biological picture. This multi-omics integration not only helps us understand the spatial distribution of gene expression, but also reveals the interactions and regulation of these genes within cells.
In addition, the combination of spatial omics and time-resolved technology allows researchers to observe the dynamic changes of cell state over time, which is of great significance for studying cell differentiation, tissue regeneration, and disease development.
Spatial omics technology still faces challenges, and it needs to be further improved in terms of sensitivity, accuracy, multi-dimensional omics, long-read detection, compatibility, throughput, cost, and accessibility of clinical samples in the future.
In terms of algorithms, spatial omics greatly expands the dimension of input signals by accurately locating cells, mapping everything from cell type and density to cell connection, communication, and microecology. In the past, low-resolution techniques focused more on deconvolution analysis of cell types, but with the current breakthrough in single-cell or subcellular resolution spatial omics technology, spatial omics algorithms can delve into cell types and their interrelationships at various biological scales, including molecular, cellular and subcellular, regional, and spatiotemporal levels.
Applications of Spatiotemporal Omics in Biology and Medicine
The spatiotemporal dynamics of living systems make it crucial to understand life processes to study how the spatial microenvironment affects cell types, states, and functions. Spatiotemporal omics analysis allows the precise mapping of molecular and cellular signals to specific locations in tissues, revealing how cells interact with their environment and how they dynamically change over time.
At present, spatiotemporal omics has shown important potential in the following five directions:
1. Organ structure: Spatiotemporal omics can reveal the organization and function of organs at the molecular and cellular levels. Not only does it help identify molecular signals in different tissue regions, but it also analyzes cell types, densities, and cell-to-cell interactions. For example, using spatiotemporal transcriptomics, an in-depth study of the macaque brain was carried out, and a detailed 3D map of the distribution of brain cells was drawn, revealing the regional specificity of cell density in the cerebral cortex, which is closely related to the hierarchical function of the cortex, providing new clues for understanding the complex functions of the brain. Spatiotemporal omics also plays an important role in the analysis of the structure and function of liver, heart, testicles, and plant organs.
2. Developmental and regeneration: Spatiotemporal omics allows scientists to study the dynamics of cells during development. It has been widely used in processes including embryonic development, brain, heart, and intestinal development and regeneration, revealing the spatiotemporal dynamics of these organs in the process of development and regeneration. For example, by studying mouse embryos, it is possible to trace the migration trajectories of interneurons and reveal the formation of different functional regions, providing a new perspective for understanding complex cellular interactions during development.
3. Evolution: By analyzing gene expression in different species, researchers can track new cell types and functional traits as they emerge during evolution. For example, in comparative studies of human, non-human primate, avian, and reptile brains, spatial omics techniques have revealed unique cell types and gene networks in the brains of different species, and these findings have contributed to the understanding of the complexity and higher functions of the human brain. In addition, spatiotemporal omics is also used in the study of plant evolution, revealing the evolutionary pathways of different photosynthesis mechanisms.
4. Human disease: Spatiotemporal omics has shown great potential in disease research, especially in unraveling the mechanisms of complex diseases such as cancer. By analyzing the spatial patterns of gene expression in tumor tissues, scientists can identify key molecular targets that influence tumor growth and invasion. For example, in liver cancer research, spatial omics technology has helped uncover cell types and their interactions in the tumor microenvironment, providing a basis for the development of more precise treatment strategies.
5. Disease pathology: spatiotemporal omics provides a new tool for disease diagnosis and treatment. By combining tissue structure with molecular characteristics, scientists can develop more precise diagnostic methods. For example, spatiotemporal omics can help identify biomarkers in cancer patients and optimize personalized treatment strategies. Although the widespread clinical application of this technology still faces challenges, such as cost and technical complexity, it paves the way for the future development of precision medicine.
 Spatiotemporal omics is used to elucidate the structure and function of organs
Spatiotemporal omics is used to elucidate the structure and function of organs
Big Science Projects on Spatiotemporal Omics
Since the completion of the Human Genome Project, large-scale genomic research has evolved from initial genome decoding (such as the International HapMap Project and the 1000 Genomes Project) to functional genomics analysis (such as the Encyclopedia of DNA Elements Project, ENCODE, and the Human Protein Atlas Project, HPA), and finally entered a new era of cellular genomics. This shift has allowed scientists to explore life processes more deeply at the cellular level.
The Human Cell Atlas (HCA) is the first international collaborative research project to map all cell types, from development to adulthood and old age. Scientists around the world have made significant progress by analyzing tens of millions of cells from nearly 10,000 individuals. On this basis, the Human Biomolecular Atlas Project (HuBMAP) is dedicated to constructing detailed biomolecular maps of human tissues to integrate cellular atlas data.
The SpatioTemporal Omics Consortium (STOC) - an open, collaborative research initiative established in 2022 to unite, organize, advance, and share global scientific efforts in spatiotemporal cellular genomics - focuses on understanding biological problems in spatiotemporal dynamics, aiming to accelerate our understanding of the biological processes of organ function, development, aging, and evolution in four directions: organ mapping, development and aging, human disease, and life evolution through large-scale spatial multiomics analysis.
In the field of brain science and oncology research, large-scale collaborative research has achieved remarkable results. For example, the BRAIN Initiative Cell Atlas Network (BICAN) is establishing a reference brain cell atlas that provides an important molecular and anatomical basis for studying brain function and disease. In oncology research, the Human Tumor Atlas Network (HTAN) is mapping the spatial and temporal transformation of tumors at single-cell resolution, which will help improve cancer detection, prevention, and treatment.
What’s next?
The advent and rapid development of spatiotemporal omics, coupled with advances in sequencing and single-cell omics, will allow us to create a "Google map" of the human body at cellular resolution in the temporal and spatial dimensions. This will enhance our understanding of the cellular and molecular basis of biological processes. At the same time, rapid advances in artificial intelligence and computational biology will significantly drive the integration of cell mapping data, imaging and clinical phenotyping data with AI algorithms. This integration will greatly advance the methods of diagnosis, treatment, and prognosis of diseases, and accelerate the clinical application of precision medicine.
This review can be accessed here: https://www.cell.com/cell/fulltext/S0092-8674(24)00834-1.



