On February 3, a research team from BGI-Research published a new method in Cell Systems. The paper introduces SpaTrack, a new computational tool for constructing spatiotemporal cell differentiation trajectories. SpaTrack combines single-cell spatial transcriptomics with optimal transport theory, offering a comprehensive framework to elucidate how cells evolve, migrate, and specialize within the dynamic contexts of tissue development, organ regeneration, and cancer progression. This innovative method leverages high-resolution data from BGI’s proprietary Stereo-seq technology, capturing both transcriptional profiles and spatial locations of individual cells in tissue samples.
 The study “Inferring cell trajectories of spatial transcriptomics via optimal transport analysis” was published in Cell Systems.
The study “Inferring cell trajectories of spatial transcriptomics via optimal transport analysis” was published in Cell Systems.
Optimal transport (OT) is a method that finds a least-cost coupling between distributions and provides intuitive quantifications of their distance. Based on this approach, SpaTrack revolutionizes traditional approaches by incorporating spatial distance into the cost measure of cell transition probabilities, moving beyond reliance on gene expression alone.
This algorithm finely tunes the balance between transcriptomic similarity and physical proximity by automatically adjusting the weights of expression and location data based on their spatial autocorrelation. Such integration allows for the reconstruction of detailed and potentially discontinuous differentiation pathways across varied tissue types.
SpaTrack not only constructs detailed trajectories from individual samples but also effectively tracks cell transitions over time, offering insights into the temporal dynamics of development and disease progression. Crucially, the tool also identifies potential transcriptional regulators, modeling the impact of transcription factors on gene expression changes throughout differentiation stages.
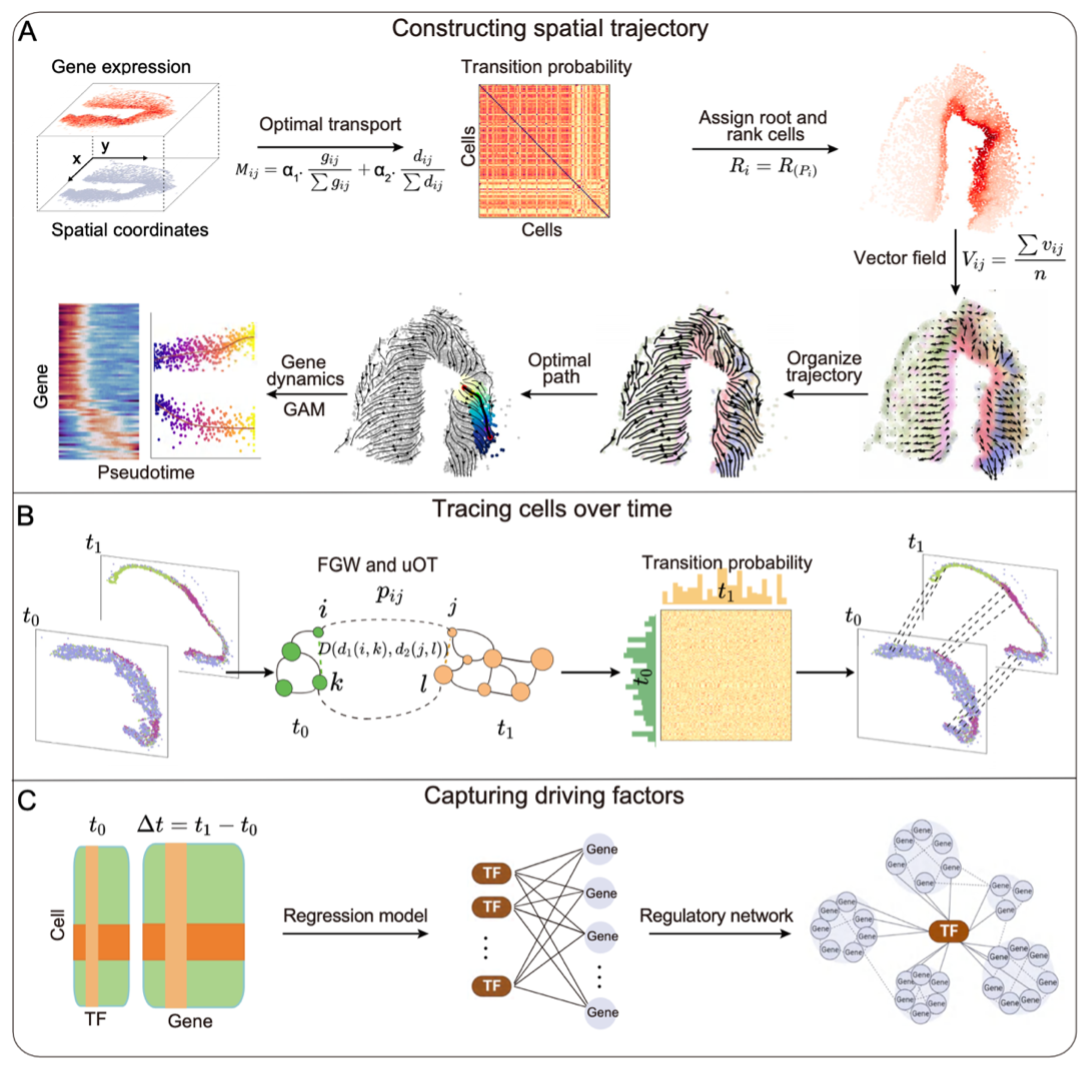 Schematic representation of SpaTrack’s functionality and key principles.
Schematic representation of SpaTrack’s functionality and key principles.
In rigorous evaluations using both simulated and real-world datasets, SpaTrack consistently outperformed existing methodologies or complemented them effectively. The research team has successfully applied SpaTrack across diverse biological systems, demonstrating its capability to illuminate complex biological processes: it has mapped out regeneration in the axolotl telencephalon, traced the developmental trajectory of the midbrain in mouse embryos, and detailed the expansion and metastatic routes in tumors. These applications highlight SpaTrack's precision in reconstructing cellular differentiation dynamics, pinpointing critical transitional states, and identifying key regulatory elements driving these processes.
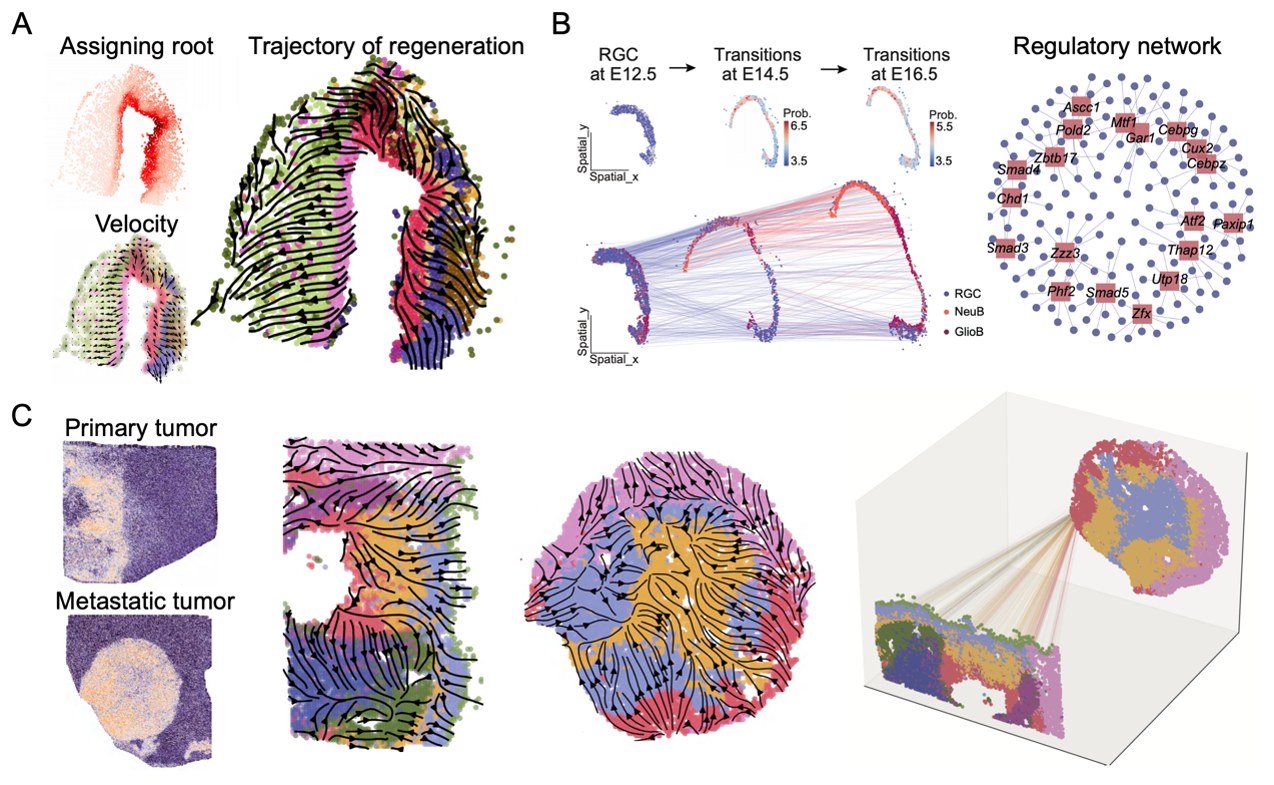 Applications of SpaTrack in elucidating axolotl brain regeneration (A), mouse embryonic midbrain development (B), and tumor growth and metastasis (C).
Applications of SpaTrack in elucidating axolotl brain regeneration (A), mouse embryonic midbrain development (B), and tumor growth and metastasis (C).
SpaTrack is now freely available as open-source software on GitHub (https://github.com/yzf072/SpaTrack), complete with detailed installation and user guides (https://SpaTrack.readthedocs.io/en/latest/index.html). Additionally, it is supported on BGI’s STOmics Cloud platform, enhancing the accessibility of spatiotemporal analytics for the global research community.
This study can be accessed here: https://www.cell.com/cms/10.1016/j.cels.2025.101194/attachment/cae808d9-0803-46cc-8071-90d5573f3ba3/mmc2.pdf



