Preeclampsia, a life-threatening pregnancy disorder, can occur after the 20th week of pregnancy or after giving birth (known as postpartum preeclampsia), affecting 2% to 4% of pregnant women worldwide. It leads to approximately 46,000 maternal deaths and about 500,000 fetal and newborn deaths each year, with preeclampsia and its associated complications causing 10 to 15 percent of maternal deaths globally.
The heterogeneity and complexity of preeclampsia make it difficult to adequately predict, treat, or prevent. However, a recent study published in the American Journal of Obstetrics and Gynecology, led by BGI Group, has shed light on a potential diagnostic method for this devastating disease.
 Noninvasive preeclampsia prediction using plasma cell–free RNA signatures published in the American Journal of Obstetrics and Gynecology.
Noninvasive preeclampsia prediction using plasma cell–free RNA signatures published in the American Journal of Obstetrics and Gynecology.
The research team employed a novel cell-free RNA sequencing method to analyze the characteristics of cell-free RNA in 715 healthy pregnancies and 202 pregnancies affected by preeclampsia before the onset of symptoms. By exploring differences in RNA biotypes between healthy and preeclampsia samples, the team developed a prediction method for preterm and early-onset preeclampsia using machine learning and validated its performance.
Cell-free RNA (cfRNA) refers to RNA that exists in body fluids, including messenger RNA (mRNA), long non-coding RNA (lncRNA), microRNA (miRNA), and others. cfRNA in plasma carries unique information from human tissues, specifically transcript fragments from various cell types. It serves as an important biomarker that can be helpful to non-invasive monitoring of maternal, placental, and fetal dynamics during pregnancy. Previous studies have also demonstrated its potential to reflect the systemic response to growing tumors and provide information about the specific tissue of tumor origin, categorized by cancer type.
Prior to the onset of preeclampsia symptoms, the research team identified 77 differentially expressed genes that could distinguish early-onset preeclampsia from healthy samples. Several of these genes have previously been reported to play significant roles in the pathological processes of preeclampsia.
The study developed separate predictive models for preterm and early-onset preeclampsia. By incorporating clinical features, both models performed well during the validation process, surpassing similar methods previously reported.
Furthermore, the study observed the down-regulation of 17 miRNAs in pregnancies with preterm and early-onset preeclampsia. Through target gene prediction of these miRNAs, the researchers discovered that six of them directly targeted four genes. Interestingly, eight out of the ten genes were utilized in both predictive models, and the expression trends of these genes were consistent with existing studies. These findings suggest that miRNAs may play a crucial role in regulating the expression of target genes associated with preeclampsia.
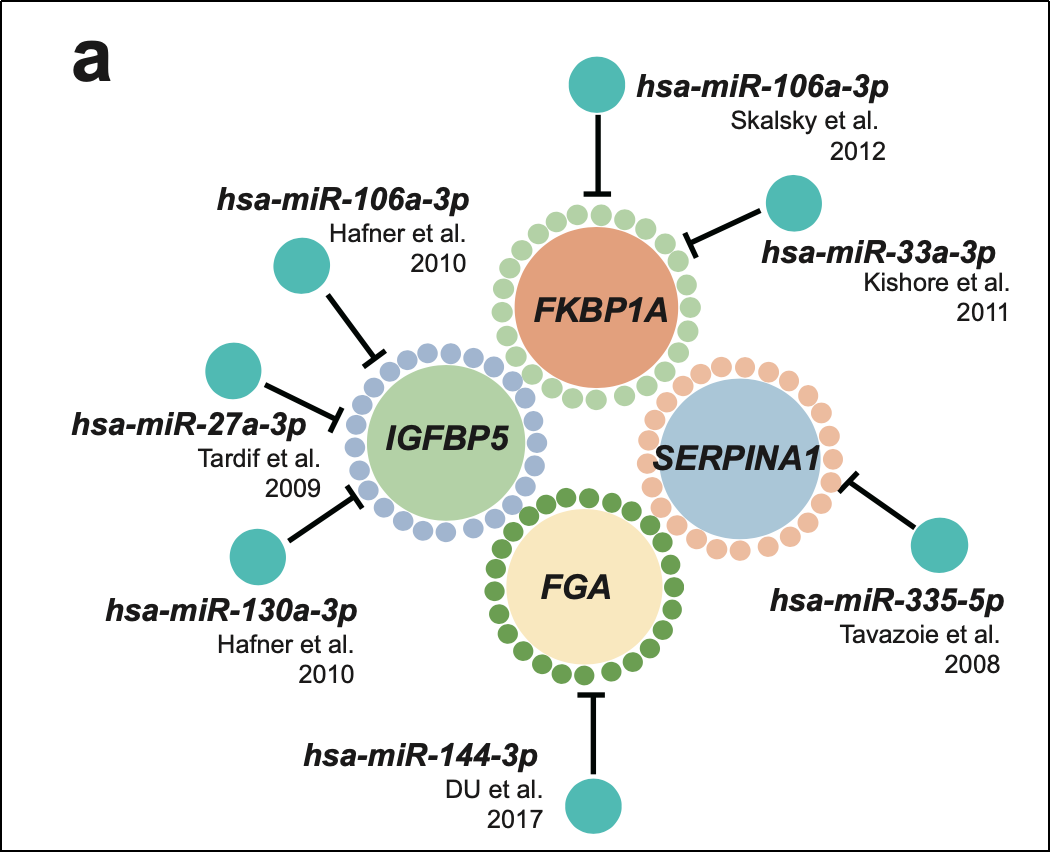 Through target gene prediction of 17 miRNAs, the researchers discovered that 6 of them directly targeted 4 genes.
Through target gene prediction of 17 miRNAs, the researchers discovered that 6 of them directly targeted 4 genes.
This study demonstrated that mRNA, microRNA, and lncRNA can serve as potential biomarkers of preeclampsia simultaneously, offering promise for the prevention of preeclampsia in the future. Abnormal changes in cell-free mRNA, microRNA, and lncRNA may help elucidate the underlying causes of preeclampsia and lead to the discovery of new therapeutic pathways to effectively reduce pregnancy complications and fetal morbidity.
The project underwent ethical review and adheres to regulations, and privacy protocols.
Read the article: https://www.sciencedirect.com/science/article/pii/S000293782300323X



