Chickpea (Cicer arietinum L.) is an important legume crop cultivated predominantly in arid and semiarid regions worldwide with global production exceeding 17 million tons annually. In addition to its high economic significance, chickpea has a high nutritional value and contributes to soil fertility by fixing atmospheric nitrogen. However, chickpea has limited genetic diversity due to several bottlenecks in its evolution, resulting in insufficient ability to resist biotic or abiotic stress which limits the breeding improvement process of the species.
 The study “Cicer super-pangenome provides insights into species evolution and agronomic trait loci for crop improvement in chickpea” was published in Nature Genetics.
The study “Cicer super-pangenome provides insights into species evolution and agronomic trait loci for crop improvement in chickpea” was published in Nature Genetics.
On May 23, a landmark study led by BGI-Research, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), Murdoch University and others, was published in Nature Genetics: “Cicer super-pangenome provides insights into species evolution and agronomic trait loci for crop improvement in chickpea”. This research marks a significant breakthrough in the effort to enhance chickpea cultivation, and presents the chickpea’s super-pangenome based on the genome assemblies of eight annual chickpea wild species and two previously reported cultivated genomes, providing unprecedented insights into the evolution of this vital legume species and providing potential for crop improvement.
The super-pangenome is the approach of developing a pangenome of the pangenomes of different species for a given genus, which provides a complete genomic variation repertoire of a genus and offers unprecedented opportunities for crop improvement.
The research team sequenced eight wild chickpea species which produced an average 143.03 Gb sequencing data per species and a total of 1.14 Tb data was generated. Additionally, they used the high-throughput chromosome conformation capture sequencing (Hi-C) data to generate chromosome-length assemblies. The analysis revealed extensive reorganization of many genome segments between cultivated and wild chickpea species. And it also demonstrates that the chickpea has three main gene pools and inferred the divergence times between the pools.
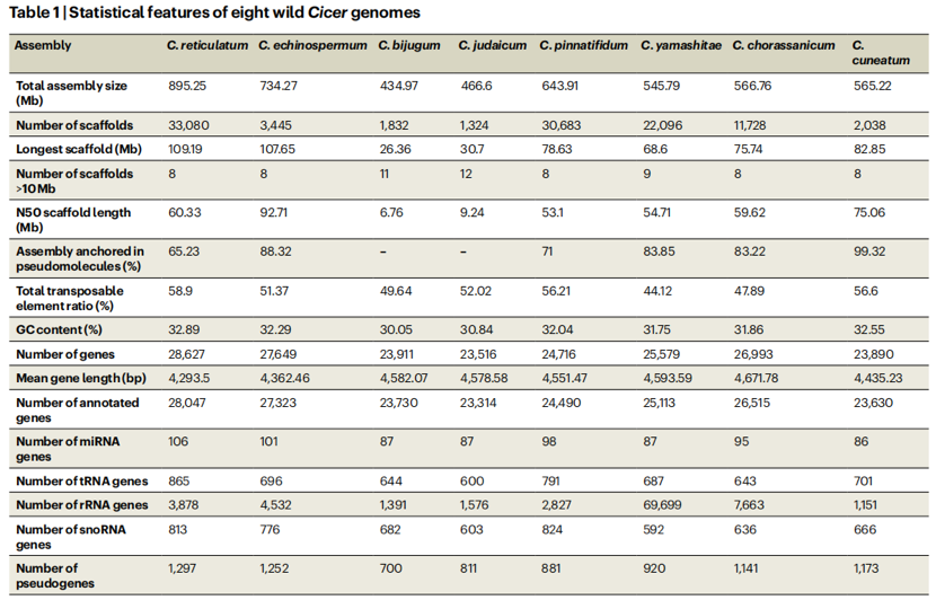 Statistical information of the genomes of eight annual wild species of the genus Cicer.
Statistical information of the genomes of eight annual wild species of the genus Cicer.
The chickpea super-pangenome analysis revealed several important findings regarding core and dispensable genes. The genes of the super-pangenome from all ten chickpea species were clustered into 24,827 gene families. Based on these gene families, the chickpea’s super-pangenome is composed of 14,748 core (present in all species), 2,958 softcore (present in nine species), 6,212 dispensable (present in two to eight species) and 909 species-specific gene families. While core gene families are involved in fundamental biological functions such as growth, DNA repair, cell communication, transport, methylation, RNA processing, metabolism, and stimuli detection, dispensable gene families are enriched for genes related to defense response, auxin response, response to biotic stimulus, cell wall modification and ion homeostasis.
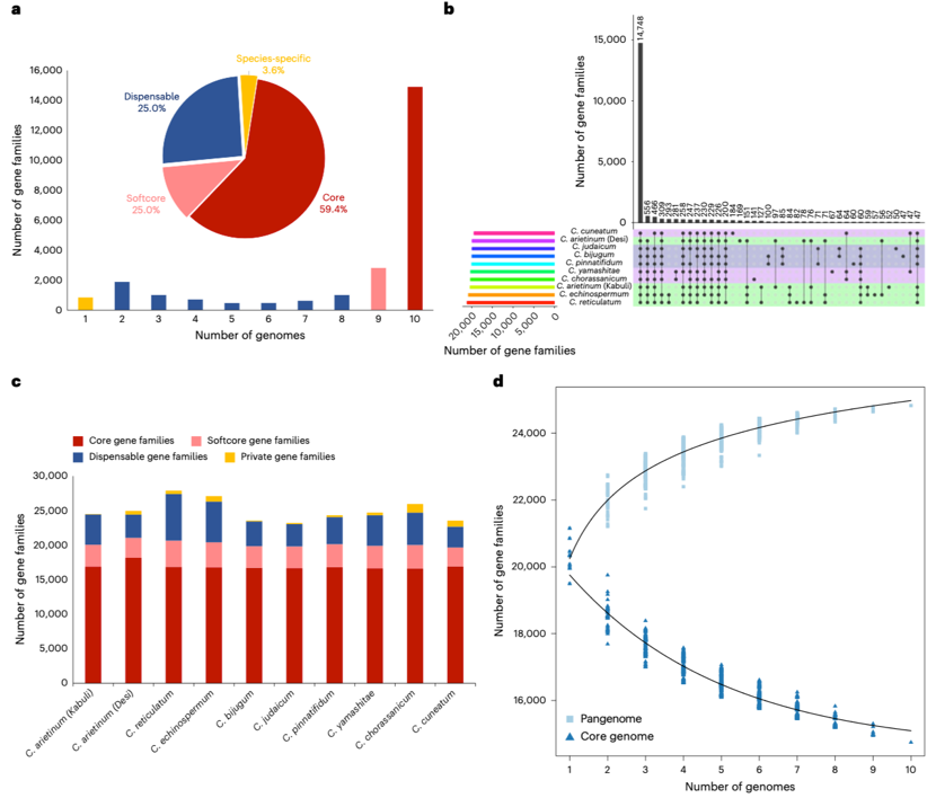 Landscape of the Cicer super-pangenome.
Landscape of the Cicer super-pangenome.
The research team identified 29.44 million nonredundant genetic variants and 6.33 million small insertions deletions. A total of 491,937 structural variations were also identified. Based on these data, a graph-based super-pangenome was constructed. The graph reveals accurate structural variation in the 477 cultivated and wild species accessions, revealing their vital role in chickpea’s flowering time, vernalization, disease resistance and other characters.
Professor Rajeev Varshney FRS, co-correspondent author and the Director of the State Agricultural Biotechnology Centre at Murdoch University, said: “The genomic resources and unique genes presented in distant relatives of modern-day chickpeas in this new study will greatly benefit chickpea breeding and the advancement of the research community in this area in Australia and globally."
“By leveraging the genetic diversity found in wild relatives of chickpea, we can significantly broaden the genetic base of cultivated chickpeas,” adds Aamir W. Khan, first author and senior scientific officer at ICRISAT. “This paves the way for breeding programs aimed at developing more robust and high-yielding chickpea varieties. Our work highlights the untapped potential of wild species to enhance the genetic toolkit available for crop improvement.”
 Since 2013, BGI has collaborated with Rajeev Varshney's team on a series of chickpea genome studies and landmark achievements (photo courtesy of Rajeev Varshney).
Since 2013, BGI has collaborated with Rajeev Varshney's team on a series of chickpea genome studies and landmark achievements (photo courtesy of Rajeev Varshney).
Since the chickpea genome project, which began in 2013, completed the first reference genome of chickpea, BGI and Professor Rajeev Varshney's team have collaborated on numerous genomic research projects and achieved significant milestones.
"It has been pleasuring to collaborate with BGI and other partners to develop the Cicer super-pangenome,” said Professor Varshney. “Our study found that the wild species have more genetic diversity and variations that could be useful for improving chickpea traits such as disease resistance, flowering time, and stress tolerance.”
Xin Liu, co-correspondent author and a researcher of BGI-Research, concluded that “Since 2013, more than a decade of scientific cooperation and research output has highlighted the value and significance of global collaboration. A series of genomics studies will promote improvements in chickpea breeding while providing solutions for enhancing other crops, addressing climate change, and ensuring food security.”
The study can be accessed here: https://www.nature.com/articles/s41588-024-01760-4



