A research team, including scientists from BGI-Research, School of Life Sciences at Zhengzhou University, and the Center for Excellence in Brain Science and Intelligence Technology and Shenzhen Institute of Advanced Technology of the Chinese Academy of Sciences, has created a comprehensive evolutionary single cell atlas of the amniote brain. Published in Developmental Cell titled "Genomic evolution reshapes cell-type diversification in the amniote brain," the study combines BGI's single-cell platform, Stereo-seq spatial transcriptomics, and long-read sequencing to map how genome evolution has reshaped brain cell diversity across reptiles, birds, and mammals over 320 million years. This work reveals key molecular mechanisms driving brain cell diversification and provides a framework for understanding brain structure and functional evolution across species.
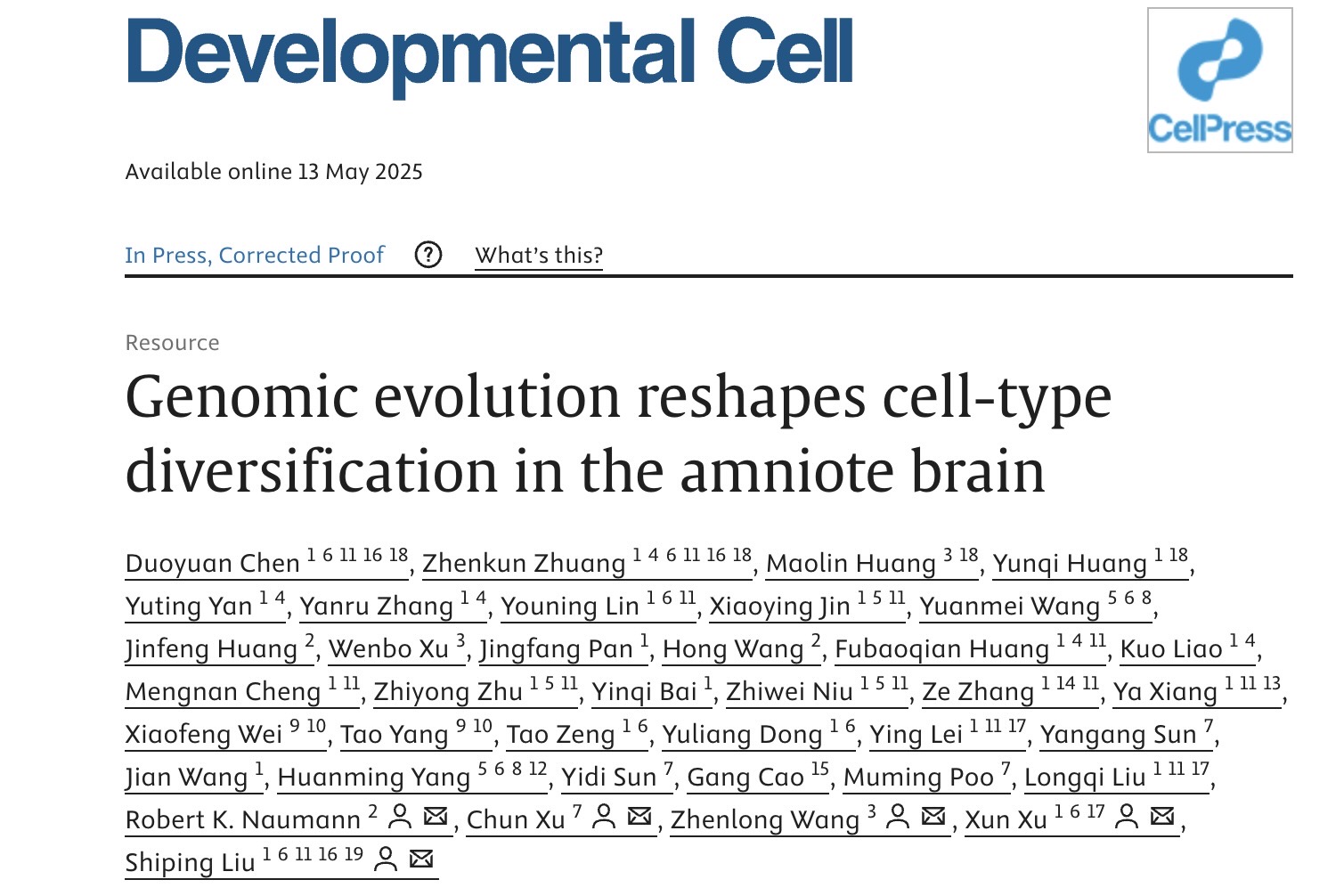 The publication in Developmental Cell distills how 320 million years of genomic tinkering through gene duplication, motif loss and paralog switching have sculpted entirely new brain-cell repertoires across reptiles, birds and mammals.
The publication in Developmental Cell distills how 320 million years of genomic tinkering through gene duplication, motif loss and paralog switching have sculpted entirely new brain-cell repertoires across reptiles, birds and mammals.
The research leverages a comprehensive single-cell atlas comprising over 1.3 million cells spanning the telencephalon and cerebellum of five key amniote species: Chinese softshell turtle, zebra finch, pigeon, mouse, and macaque. A significant portion of the data, particularly from non-mammalian species, was generated using BGI's high-throughput single-nucleus RNA sequencing platform. Cross-species comparisons were strengthened by enhanced gene annotations, especially for less-characterized species like turtles and pigeons, facilitated by BGI's long-read sequencing technology. BGI's Stereo-seq spatial transcriptomics further validated these findings by mapping in-situ gene expression patterns and localizing cell types within the brain tissue architecture.
This integration of multiple omics approaches, powered by an entropy-optimized graph convolutional network model (entropy-scGCN) for reliable cross-species cell type mapping, enabled researchers to resolve the evolutionary trajectories of amniote brain cell types with unprecedented spatiotemporal detail.
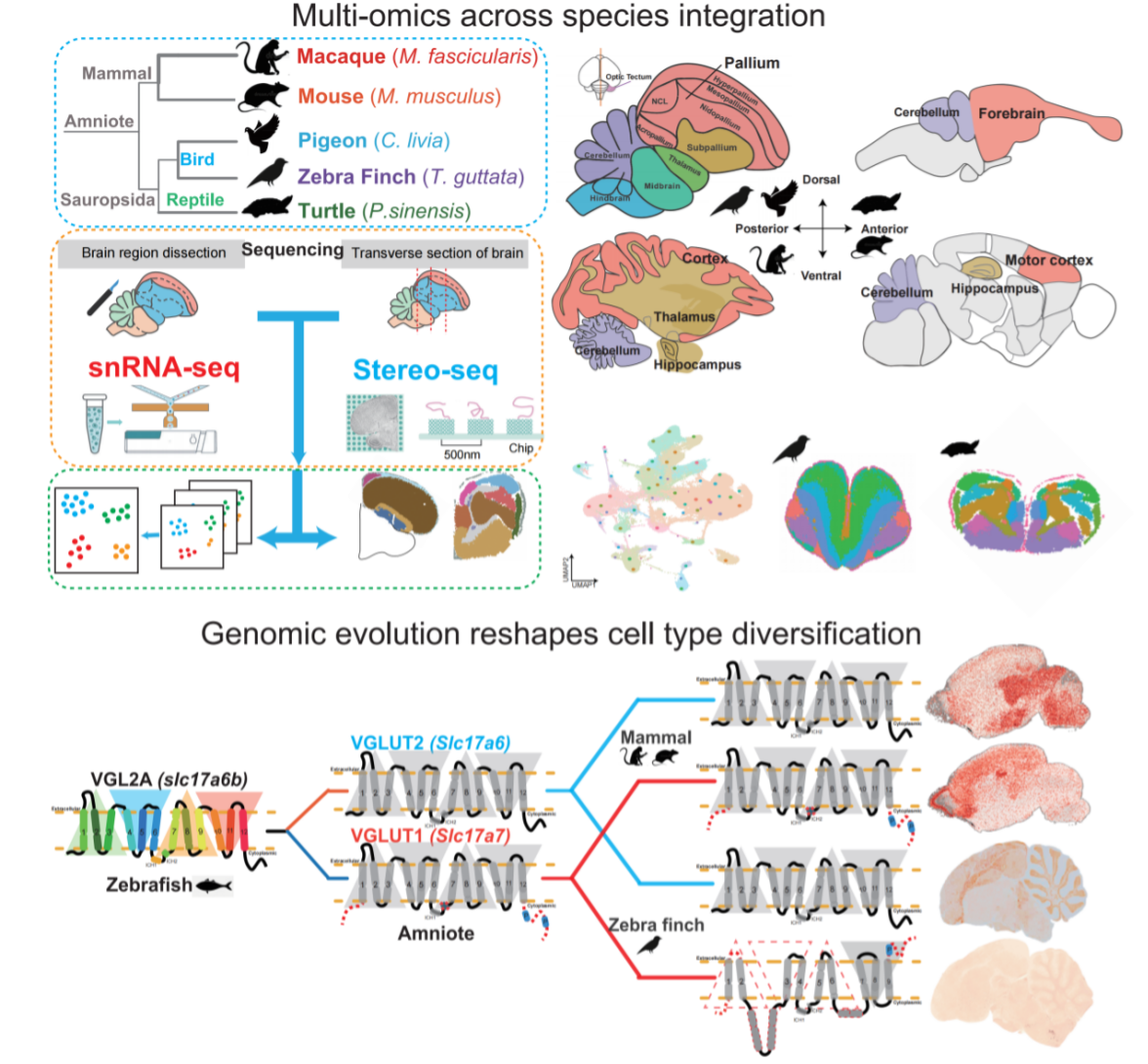 Main Results and Core Discoveries highlights the study’s most important findings from single-cell and spatial multi-omics, showing how gene duplications and paralog shifts reshape brain cell repertoires.
Main Results and Core Discoveries highlights the study’s most important findings from single-cell and spatial multi-omics, showing how gene duplications and paralog shifts reshape brain cell repertoires.
Among the key revelations is the striking divergence in molecular mechanisms of excitatory neurons between birds and mammals. The study shows that avian excitatory neurons primarily express the gene SLC17A6 (encoding VGLUT2), across the whole brain regions, while mammalian neocortical neurons have specialized to express its paralog, SLC17A7 (encoding VGLUT1), in most of the cortex regions.
In mammals, while SLC17A6 expression persists in other brain regions, including ancient cortical areas like the thalamus, the earlier diverging turtles exhibit widespread expression of both SLC17A6 and SLC17A7. This contrasting pattern of gene usage likely results from the functional loss of SLC17A7 in the avian lineage, a conclusion bolstered by genomic analyses demonstrating that even subtle mutations within crucial protein domains can induce substantial structural changes. These molecular observations shed light on the diverse strategies for neurotransmitter release and processing efficiency that have evolved, reflecting the distinct brain functional adaptations across these key amniote groups.
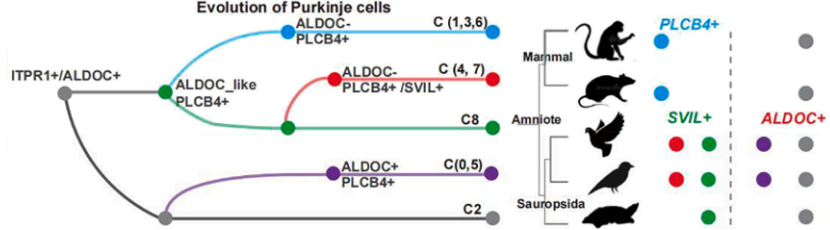 Inferred Evolutionary Inferred Evolutionary Model of Cerebellar Purkinje Cell Subtypes traces how an ancestral Purkinje population gave rise to mammal-specific PLCB4+ and bird-specific SVIL+ lineages, revealing independent adaptations in motor learning and circadian regulation. It reframes our view of cerebellar diversity across vertebrates.
Inferred Evolutionary Inferred Evolutionary Model of Cerebellar Purkinje Cell Subtypes traces how an ancestral Purkinje population gave rise to mammal-specific PLCB4+ and bird-specific SVIL+ lineages, revealing independent adaptations in motor learning and circadian regulation. It reframes our view of cerebellar diversity across vertebrates.
Highlighting the unique evolutionary adaptations within the avian lineage, this study identified a novel Purkinje cell subtype in the bird cerebellum, termed SVIL+ cells, demonstrating evolutionary innovation even in conserved brain regions. The gene expression signature of these SVIL+ cells is markedly distinct from that of mammalian ALDOC+ and PLCB4+ Purkinje cells, showing an enrichment in genes linked to learning, memory, and circadian rhythm regulation. Strikingly, this avian-specific cell type exhibits a high number of positively selected genes, suggesting adaptive evolution of cerebellar functions crucial for birds, potentially underpinning their sophisticated behaviors like flight and complex vocalizations. Analysis of regulatory networks further revealed the specialized gene expression program of SVIL+ cells, underscoring their functional specialization shaped by the unique ecological demands faced by birds.
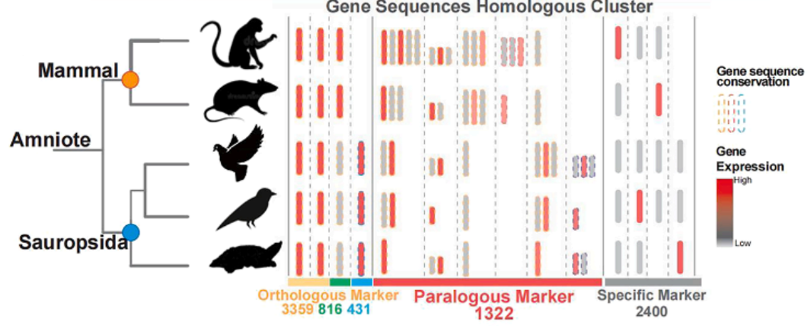 Statistics Linking Amniote Gene-Family Evolution to Brain Cell Expression Features quantifies thousands of orthologues, paralogs and species-specific genes across cell types, showing that gene-family expansion fuels neural innovation and enables mapping from genotype to cell function.
Statistics Linking Amniote Gene-Family Evolution to Brain Cell Expression Features quantifies thousands of orthologues, paralogs and species-specific genes across cell types, showing that gene-family expansion fuels neural innovation and enables mapping from genotype to cell function.
To elucidate the genomic underpinnings of cellular diversity, the researchers discovered that gene family evolution has profoundly sculpted brain cell type regulatory networks across amniotes. Notably, 20-35% of differentially expressed genes in neurons can be attributed to gene family expansion and duplication events in ancestral amniotes, revealing a "dual-track" evolutionary mechanism. This mechanism posits that conserved transcription factors maintain core regulatory programs, while species-specific cellular diversification arises through processes such as paralog sub-functionalization. Ultimately, this "dual-track" evolutionary strategy explains how ancient genomic events laid the groundwork for the emergence of diverse cell type specializations and the subsequent development of intricate neural circuits.
Transcending a simple catalog of cell types, this landmark study establishes an integrated molecular, cellular, and spatial framework for probing brain evolution across vast evolutionary distances. The comprehensive and openly accessible datasets and analytical tools, hosted at the China National GeneBank DataBase (CNGBdb) via https://db.cngb.org/cdcp/absna/, provide an essential resource for future research into brain complexity and adaptation. Looking ahead, the researchers aim to inspire the application of this approach across the tree of life, ultimately striving to reconstruct the evolutionary origins of higher cognition and to elucidate the fundamental biological principles underlying the emergence of intelligence.
This study can be accessed here: https://doi.org/10.1016/j.devcel.2025.04.014



