When scientists examine a slice of tumor tissue under the microscope, they see only part of the story. Gene activity, cellular architecture, and the precise location of each cell weave together a complex tapestry that determines how a cancer grows, spreads, and responds to therapy. Existing software often stitches those threads together only at short range, overlooking critical long-distance connections and leaving clinicians with an incomplete picture.
In the latest issue of Briefings in Bioinformatics, researchers at BGI-Research, working with collaborators in Singapore and elsewhere in China, have debuted an artificial in-telligence platform called StereoMM that fuses all three data layers into a single, high-definition map. In proof of principle studies spanning lung, breast, and colorectal can-cers, the system flagged malignant regions invisible to traditional pathology, discov-ered prognostic gene signatures, and, most strikingly, separated colorectal cancer pa-tients into treatment-relevant subtypes with improved accuracy compared to uni-modality approaches, highlighting the potential for future advancements.
 The study “StereoMM: a graph fusion model for integrating spatial transcriptomic data and pathological images” was published in Briefings in Bioinformatics.
The study “StereoMM: a graph fusion model for integrating spatial transcriptomic data and pathological images” was published in Briefings in Bioinformatics.
StereoMM builds on BGI's proprietary Stereo-seq spatial transcriptomics technology, which captures gene expression at near single cell resolution across entire tissue sec-tions. The team combines those readouts with matching hematoxylin-and-eosin imag-es and the XY coordinates of every spot on the slide, then feeds the three data streams into a deep learning architecture that marries a cross-attention module, adapted from natural language processing, with a variational graph autoencoder. The cross-attention component teaches the model to "look" across the full field of view and weigh relationships between distant cells, while the graph autoencoder aggregates local neighborhoods to preserve spatial context. The result is a compact numeric represen-tation that encapsulates molecular programs, morphology, and geography, ready for downstream discovery or clinical decision making.
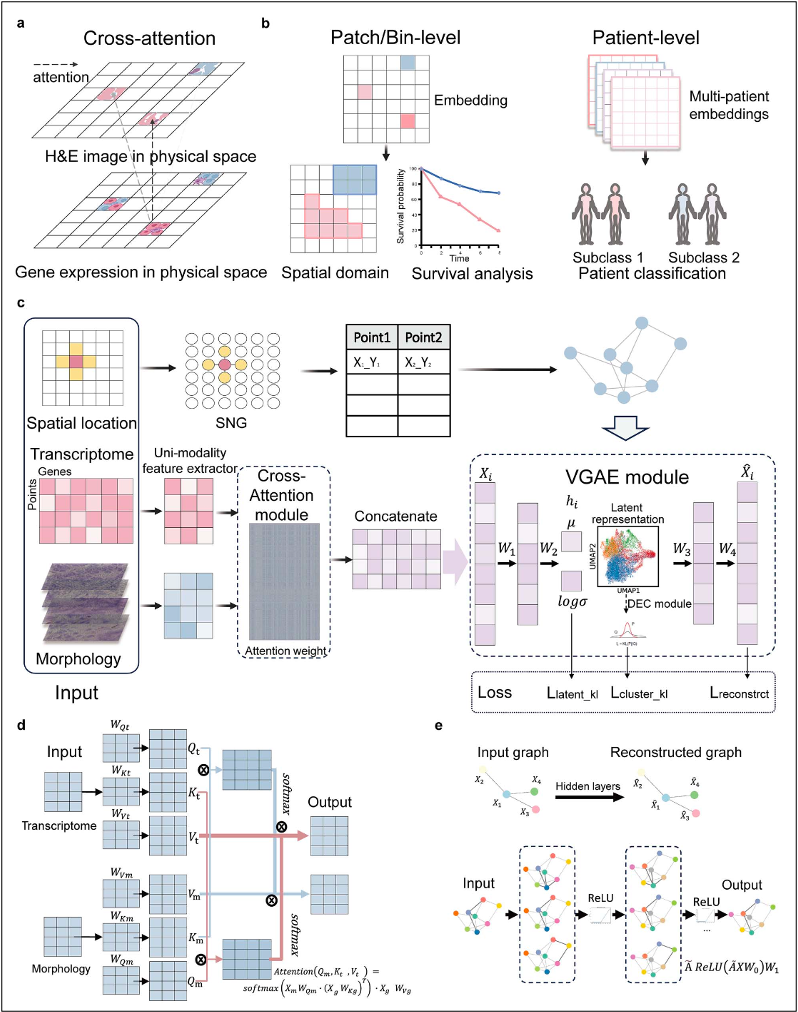 StereoMM merges spatial transcriptomics and pathology via cross-attention and graph autoencoding, producing integrated embeddings that enable accurate tumor region detection and patient-level classification.
StereoMM merges spatial transcriptomics and pathology via cross-attention and graph autoencoding, producing integrated embeddings that enable accurate tumor region detection and patient-level classification.
Applying the approach to colorectal cancer samples, the investigators showed that StereoMM cleanly divided patients into proficient and deficient mismatch-repair (pMMR and dMMR) groups, the decisive factor for prescribing immunotherapy, where previous single modality tools had stumbled. In lung adenosquamous carcinoma, the model drew boundaries that matched expert pathology more closely than eight other leading algorithms and ran faster while doing so. In a breast cancer spatial analysis, StereoMM uncovered two tumor regions with distinct stemness profiles and highlight-ed two tumor suppressor genes, SEMA3B and TFF1, whose high expression predicted better outcomes in a cohort of 334 HER2-positive patients.
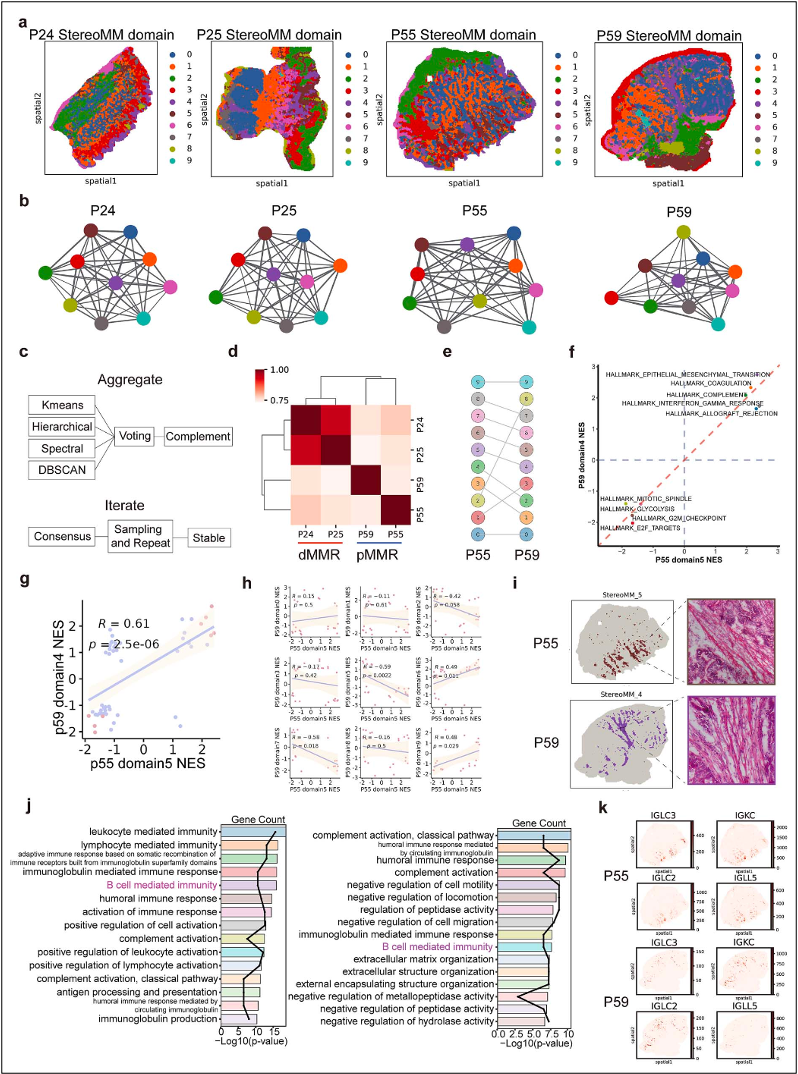 StereoMM-derived spatial domains build patient graphs that robustly separate dMMR from pMMR colorectal cancers and reveal immune driven functional differences across matched tumor regions.
StereoMM-derived spatial domains build patient graphs that robustly separate dMMR from pMMR colorectal cancers and reveal immune driven functional differences across matched tumor regions.
The research team stated that their primary goal is to convert spatial omics data into insights that clinicians can act upon. By extracting attention weights from the model, they can quantify how much each data layer (genes, images) contributes to a decision, providing oncologists with an interpretable readout rather than a black box. They not-ed that StereoMM includes adjustable hyperparameters, allowing users to tune the in-fluence of each modality to suit different tissue types or research questions.
To spur adoption, the team has released StereoMM as open-source code on GitHub and embedded it in BGI's cloud-based STOmics platform, where researchers can up-load spatial transcriptomics data and run the workflow with point and click ease. Look-ing ahead, the group plans multicenter collaborations to validate StereoMM in larger patient cohorts and to expand the framework to additional data types, including spatial proteomics and multi-slice three dimensional reconstructions.
Researchers, clinicians, and industry partners interested in exploring StereoMM can access the code at https://github.com/STOmics/StereoMMv1 and the accompanying tutorial on STOmics Cloud. Raw datasets have been deposited in the China National GeneBank DataBase (CNGBdb) under the accession numbers provided in the paper.
This research can be accessed here: https://doi.org/10.1093/bib/bbaf210



