(Continued from PART 2)
Dr. Xu Xun: Single-cell sequencing has certain limitations as it requires tissue dissociation, and when cells dissociated, they lose spatial location information. As a result, when we see a group of specific type of cells, there is no way to explain what they do and where they located. But the explanation of biological problems needs to associate with structure – it is the structure that determines function. To understand the structure, the observation of cells must be done in situ, which is why spatiotemporal omics is so important.
In the history of life sciences, the invention of the microscope started a new chapter for research as well as a turning point in the discovery journey of human being and other life forms. With the microscope, people could see the basic unit of life, cell, and further discover many different cell types. This led to the discovery of bacteria. Thanks to the findings by microscopy, drugs such as penicillin and others for bacterial infection treatment were produced, which became one of the factors that extended average human life expectancy from 30 years old to more than 70 years.
In 2001, the completion of a draft sequence of human genome marked the second turning point in the entire life sciences. Thanks to the Human Genome Project, researchers could explain the mechanism of life at the molecular level, which offers a foundation for today's precision medicine. Take COVID-19 for example. It was through genome sequencing that the world got to understand that it is caused by a type of coronavirus.
However, both methods didn’t go without limitations. The microscopic observation of life only tells morphological information but offers no molecular or mechanism-level data. DNA sequencing only gives molecular information but offers no structural information. Spatiotemporal omics solves these problems by integrating data on both structural and molecular levels at the same time - a first in human history.
We consider spatiotemporal omics technology the third inflection point in life science. BGI’s spatiotemporal omics technology, Stereo-seq, uses a sequencing chip with DNA nanoball-patterned arrays. Each nanoball is 500 nanometers in distance that allows the chip to capture the in situ mRNA information of a tissue. With the coordination data, the mRNA information can be localized to its real spatial position. Eventually, all spatial position of mRNAs of a tissue can be reveled on the molecular level. Such is the power of spatiotemporal omics that Nature Methods crowned it the “Method of the Year” for 2020.
In May 2022, together with a number of global science institutes, BGI released the world’s first panoramic atlases of life in Cell. This is the first time an ultra-high-precision analysis of gene alternation and cell development has been performed from the temporal and spatial perspectives.
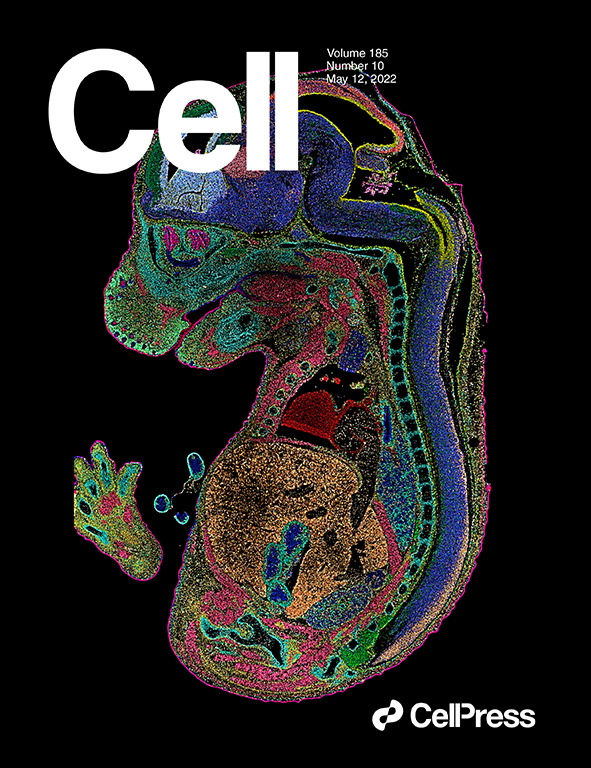 Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays published in Cell and selected as cover story.
Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays published in Cell and selected as cover story.
Among them, the atlas of mouse embryonic development was published in Cell as a cover story. Using BGI’s Stereo-seq technology, researchers revealed the organ and cell development of mouse embryo from 9.5 days to 16.5 days. This is a panoramic and ultra-high-precision dynamic description of the molecules and structures in the mouse embryonic development process, and is the most detailed atlas of the entire embryonic development process in today’s life science.
Many scientists have previously tried but failed due to the limitation of conventional technology, which cannot achieve the single-cell level resolution, nor offer a field of view to show the entire tissue, organ or even the complete individual. In contrast, BGI’s Stereo-seq is ahead of current technology in both resolution and field of view.
Stereo-seq technology is creating significant impacts in four areas that will reshape our understanding of life:
Reshape our understanding on the structure of life. Take brain research for example. In the past, scientists knew that different brain regions may perform different functions, but it was impossible to associate regions with functions with microscopic observation. With Stereo-seq, we can carry out molecular function-based partitioning of different regions of the brain. This is the first time the brain can be explained with spatiotemporal omics, which is different from the explanation using CT, MRI and other technologies.
Redefine disease. Currently, diseases are defined through two pathological techniques, the H&E (hematoxylin and eosin) staining and IHC (immunohistochemistry). The shortcoming of both conventional methods is that there is no way to digitize the process and result. In addition, both methods are heavily reliant on doctors’ experience, which can be inaccurate or wrong. Stereo-seq can provide a whole new digitized approach to redefine diseases, which is completely different.
Re-understanding development. As the mouse embryonic development research showed, Stereo-seq helps us to understand how a fertilized egg develops into a mature individual from a different perspective.
Recognizing the origin of species. We can have a completely different understanding of the origin and adaptation of species through the perspective of spatiotemporal omics.
The SpatioTemporal Omics Consortium (STOC) was established with the goal of analyzing life with spatiotemporal omics technology. STOC will promote multi-country, multi-disciplinary, and multi-field cooperation, and aims to produce blockbuster scientific results. Through the cooperation consortium, members communicate and learn from each other to reach consensus on science discoveries and promote the development of life sciences. As of June 2022, STOC has more than 100 scientists from 20 countries, including biologists, mathematicians, computer scientists and other professionals from Harvard University, Cambridge University, and Oxford University.
(To be continued in PART 4)



