On November 11, 2024, a groundbreaking study titled “Spatiotemporal Modeling of Molecular Holograms” was published in the prestigious journal Cell, led by an international collaboration between BGI-Research, Wuhan University’s School of Electronic Information, and other research institutions. This collaborative effort introduces Spateo, a groundbreaking algorithm toolkit that enables precise 3D reconstruction of whole mammalian organs and mouse embryos and systematic modeling of dynamic developmental processes, which marks a critical advancement in spatial transcriptomics and the study of life from a spatiotemporal perspective.
 The Study “Spatiotemporal Modeling of Molecular Holograms” was published in the prestigious journal Cell.
The Study “Spatiotemporal Modeling of Molecular Holograms” was published in the prestigious journal Cell.
Spateo: Unlocking the Secrets of Embryogenesis
Understanding how cells migrate and interact during early development is crucial for discovering the causes of congenital diseases. By visualizing the dynamic processes that shape tissues and organs, scientists can gain deeper insights into developmental biology. Current analytical framework for spatial omics technologies, however, often focuses on visualizing spatial expression and signals but lack the comprehensive tools for in-depth analyses of cellular changes over time.
Spateo fills this gap by offering a unified framework that allows researchers to create highly detailed 3D "molecular holograms", for example, of an entire mouse embryo. The study then generalizes the 3D alignment to predict the 4D spatiotemporal cell migrations, bridging macroscopic morphogenesis with molecular dynamics at two key stages of mouse embryonic development (E9.5 and E11.5) and of Drosophila embryonic development (Stage 11 and Stage 13). This level of detail will significantly advance our understanding of how gene expression and cellular interactions drive the formation of organs during development.
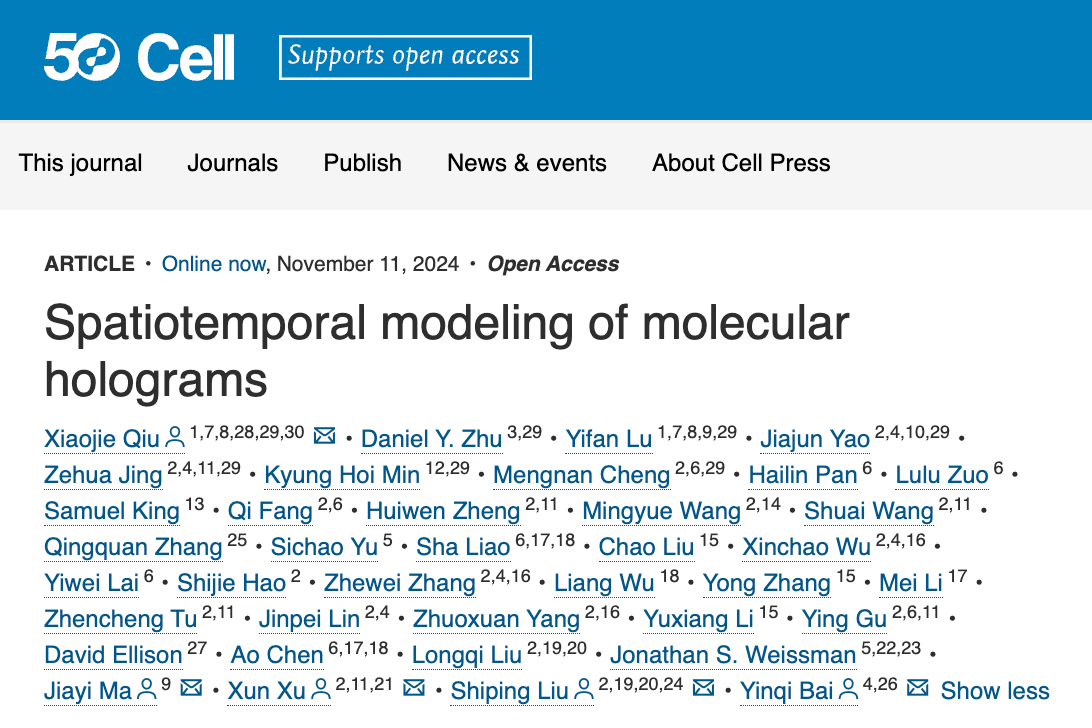
 Spateo, a versatile tool for 3D spatiotemporal transcriptomics modeling at single-cell resolution and whole-embryo scale, demonstrated on the mouse and Drosophila embryos.
Spateo, a versatile tool for 3D spatiotemporal transcriptomics modeling at single-cell resolution and whole-embryo scale, demonstrated on the mouse and Drosophila embryos.
Overcoming Challenges in 3D Alignment
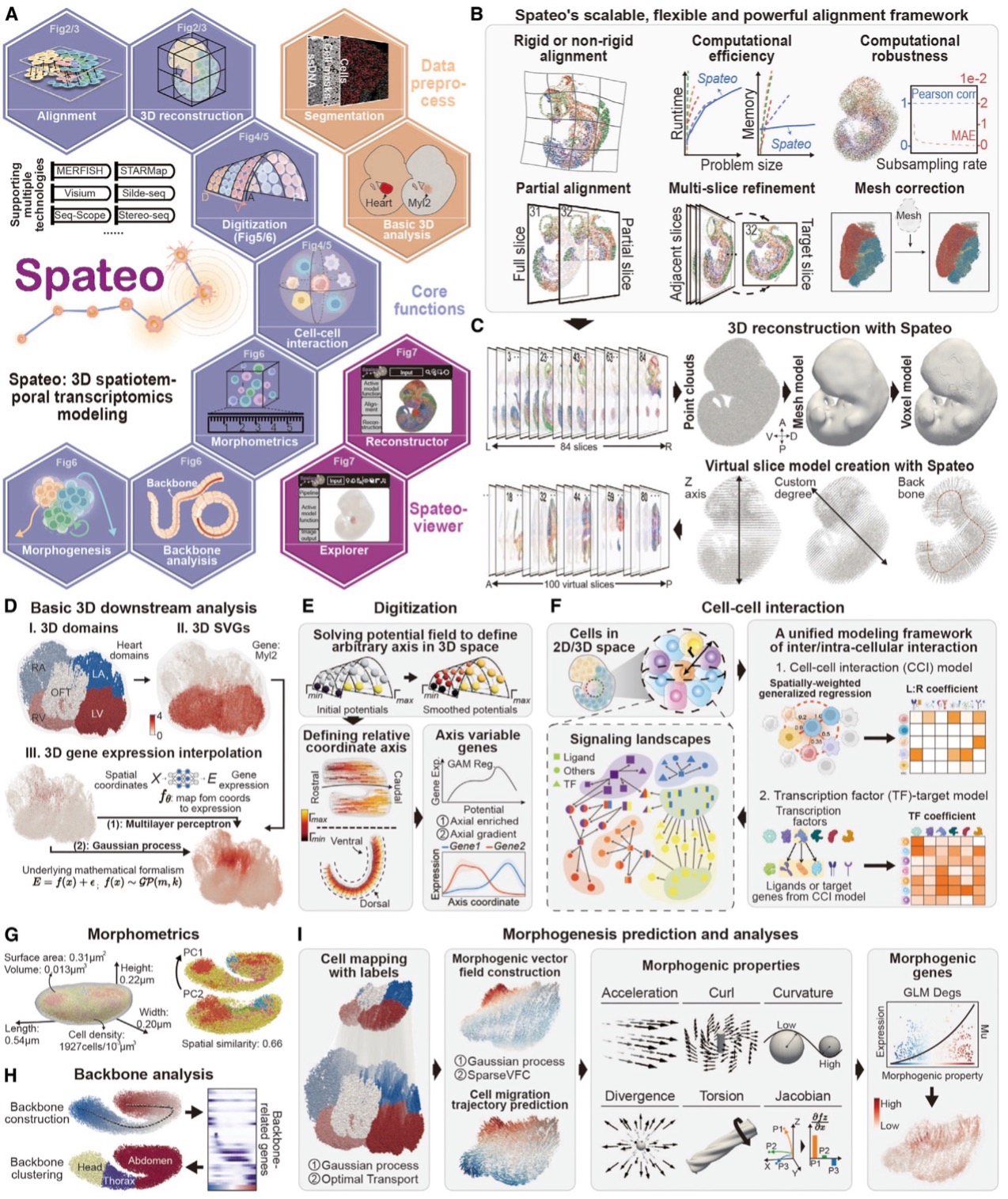
One of the major challenges in reconstructing 3D structures from tissue sections is dealing with missing regions and tissue deformations caused by sample preparation. Existing methods struggle to align large-scale embryonic sections while accounting for local distortions. Furthermore, it doesn’t allow multi-slice alignment to account for the accumulated error during sequential alignment, let alone mesh correction when an image-based 3D mesh is provided.
Spateo, however, introduces a powerful new mathematical approach that overcomes these limitations. Using pairwise alignment, multi-slice refinement, and mesh correction techniques, Spateo can align tissue sections more accurately and at a larger scale than ever before.
The algorithm's ability to handle both rigid and non-rigid transformations allows it to compensate for local distortions and missing sections, ensuring that even complex tissues can be reconstructed in 3D with high precision. This also provides an intricate view of how cells migrate and interact across key stages of development, whether on 2D or reconstructed 3D objects.
To validate Spateo’s effectiveness, the research team applied the algorithm to a variety of biological systems, including the mouse forebrain hemisphere, human metastatic lymph nodes, macaque cortex, and mouse embryo, showcasing its ability to handle complex datasets that outperform existing state-of-the-art methods. The validation results confirmed the algorithm’s accuracy in aligning consecutive tissue sections and its potential to provide new insights into tissue organization and cellular interactions in both healthy and diseased states.
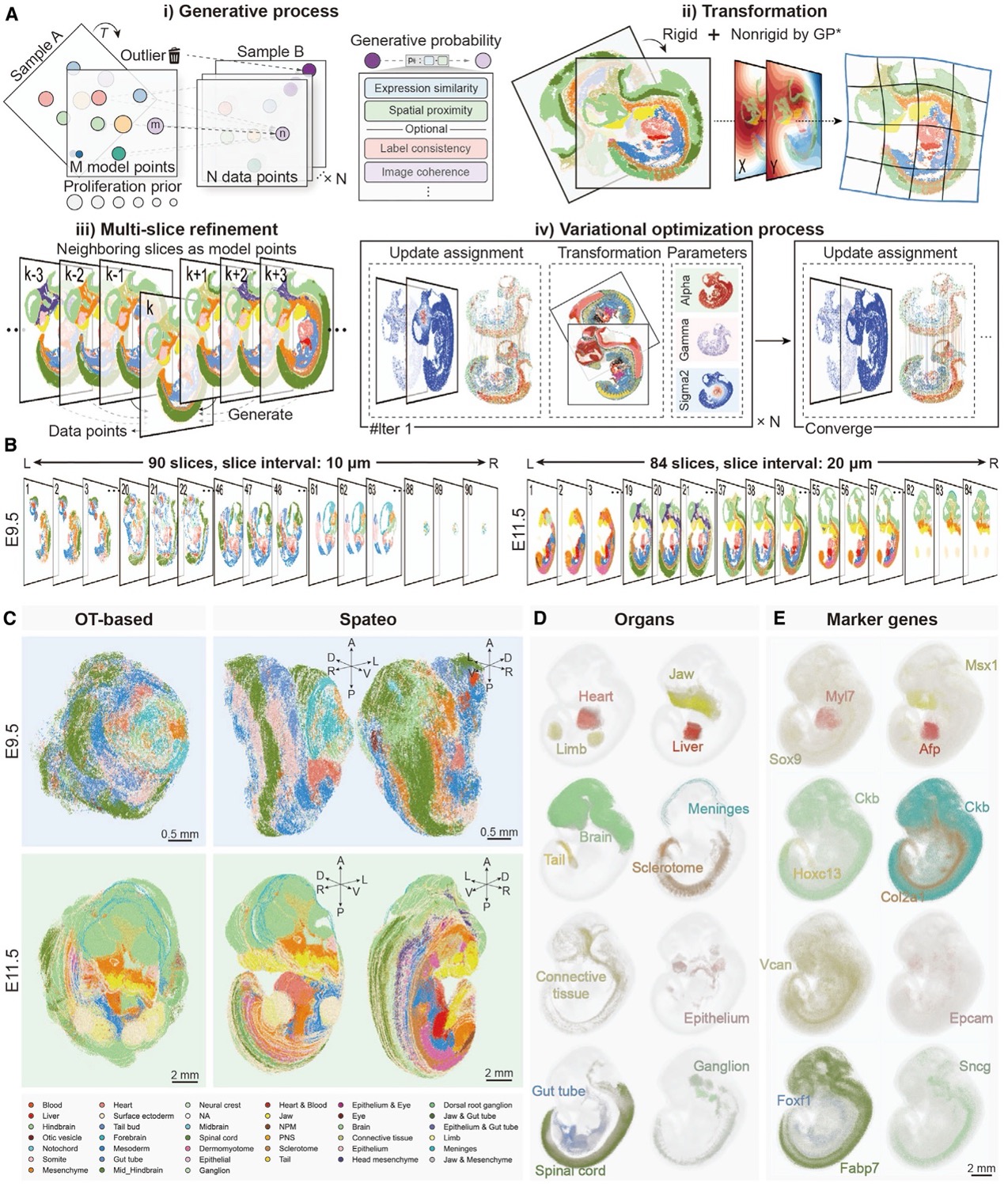 Spateo enables accurate, efficient, and scalable reconstruction of 3D molecular holograms of whole mouse embryos.
Spateo enables accurate, efficient, and scalable reconstruction of 3D molecular holograms of whole mouse embryos.
3D Digitization and Spatial Regression of Cell-Cell Interaction for Understanding Embryonic Regionalization
Embryonic regionalization is the developmental process through which an embryo is segmented into distinct regions or compartments, each destined to form specific structures or tissues. This segmentation involves a complex interplay of genetic signals, positional cues, and molecular gradients that collectively determine the fate and identity of cells within these regions.
Spateo introduces a novel digitization method based on potential functions, enabling the creation of arbitrary axes in any topologically complex 2D or 3D structure. This approach facilitates the exploration of genes with gradient dynamics and leverages a spatial regression function to infer upstream and downstream gene regulations through cell-cell interaction (CCI) inference.
Using a reconstructed 3D mouse embryo dataset, Spateo investigated emergent structures that were previously challenging to examine in detail, including the zona limitans intrathalamica (ZLI), midbrain-hindbrain boundary (MHB), and spinal cord. This method enabled the identification of specific gene expression patterns and signaling landscapes that govern cell fate in surrounding spatial regions. The digitization and CCI methods can also be applied to studies of non-small cell lung cancer, the macaque cortex with its complex topology, and even subcellular RNA distribution with high spatial resolution.
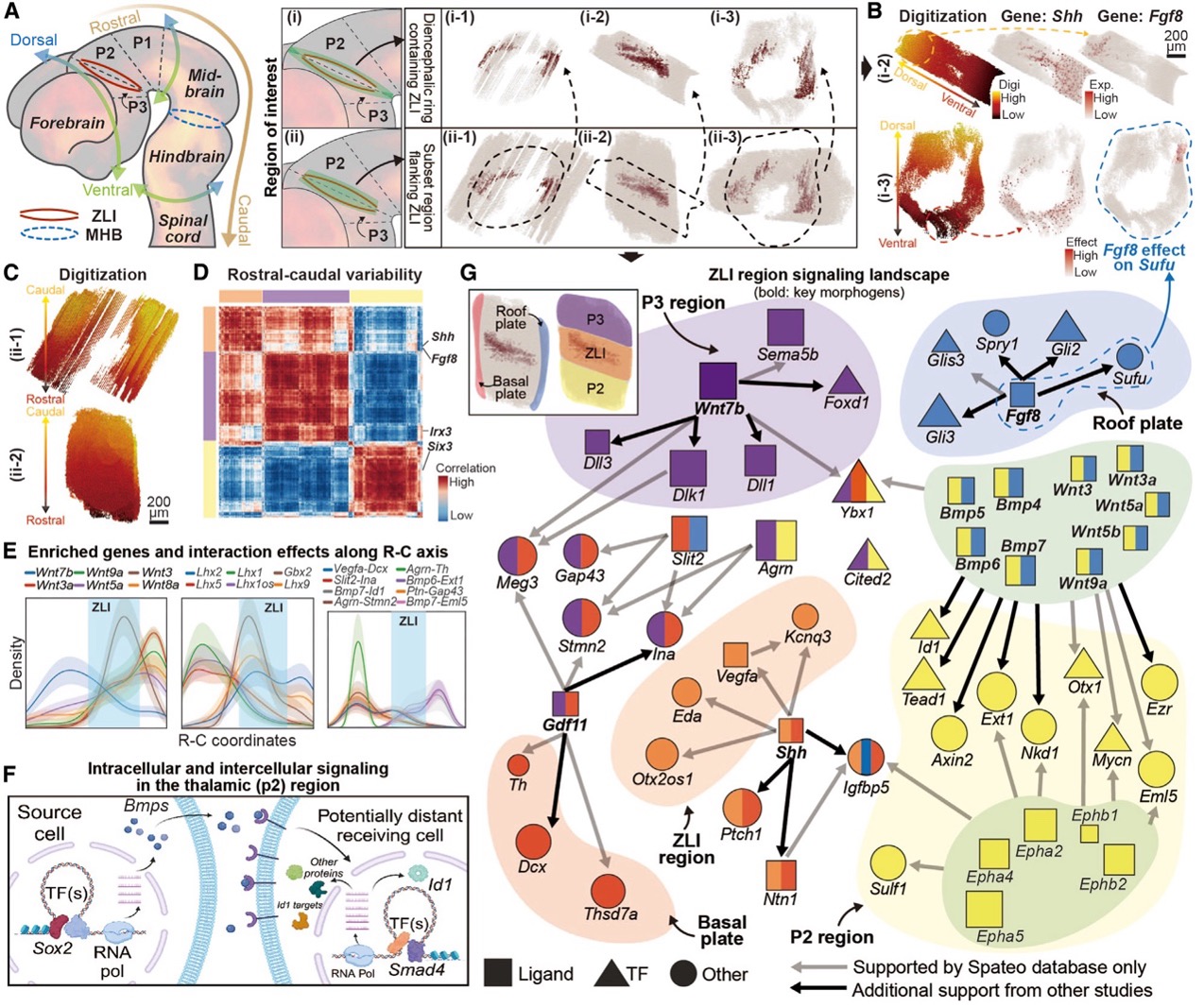 Spateo identifies networks of intercellular signaling and intracellular regulations in the developing brain proximal to the zona limitans intrathalamica.
Spateo identifies networks of intercellular signaling and intracellular regulations in the developing brain proximal to the zona limitans intrathalamica.
Highlighting Key Morphometric Factors
One of the most exciting and powerful features of Spateo is its ability to create 3D morphometric vector fields that reveal critical physical properties of tissue development, which links macroscopic cellular movements with underlying molecular pathways. These vector fields allow researchers to measure: 3D curl (the degree of rotation at any given point in the tissue); acceleration (the speed at which cells or tissues are moving); curvature (the bending of tissue structures); torsion (how much a 3D structure twists); divergence (whether the tissue is expanding or shrinking), and Jacobian (how movement along one axis affects movement along another).
These measurements are not just abstract mathematical concepts - they have real physical meanings and are crucial for understanding how tissues form and change during development. By analyzing these properties, Spateo can identify key morphogenetic genes - genes that play a critical role in shaping tissues - by finding those with significant correlations to these morphometric traits. This capability provides new avenues for studying how organs form and how genetic disruptions can lead to developmental disorders.
For example, Spateo uncovered distinct cell movement patterns that highlight the asymmetry in its formation in heart development. The right atrium (RA) demonstrated the highest acceleration, while both the right ventricle (RV) and left atrium (LA) exhibited the greatest curl, indicating significant rotational movement in these regions. In contrast, the left ventricle (LV) showed the lowest divergence, suggesting minimal expansion or contraction. These findings correspond with the developmental timeline of these heart structures, as the LV originates from the first heart field, while the RA, LA, and RV, which arise from the second heart field, are integrated into the heart at a later stage. This deeper understanding of the heart’s asymmetrical development could have significant implications for studying congenital heart defects.
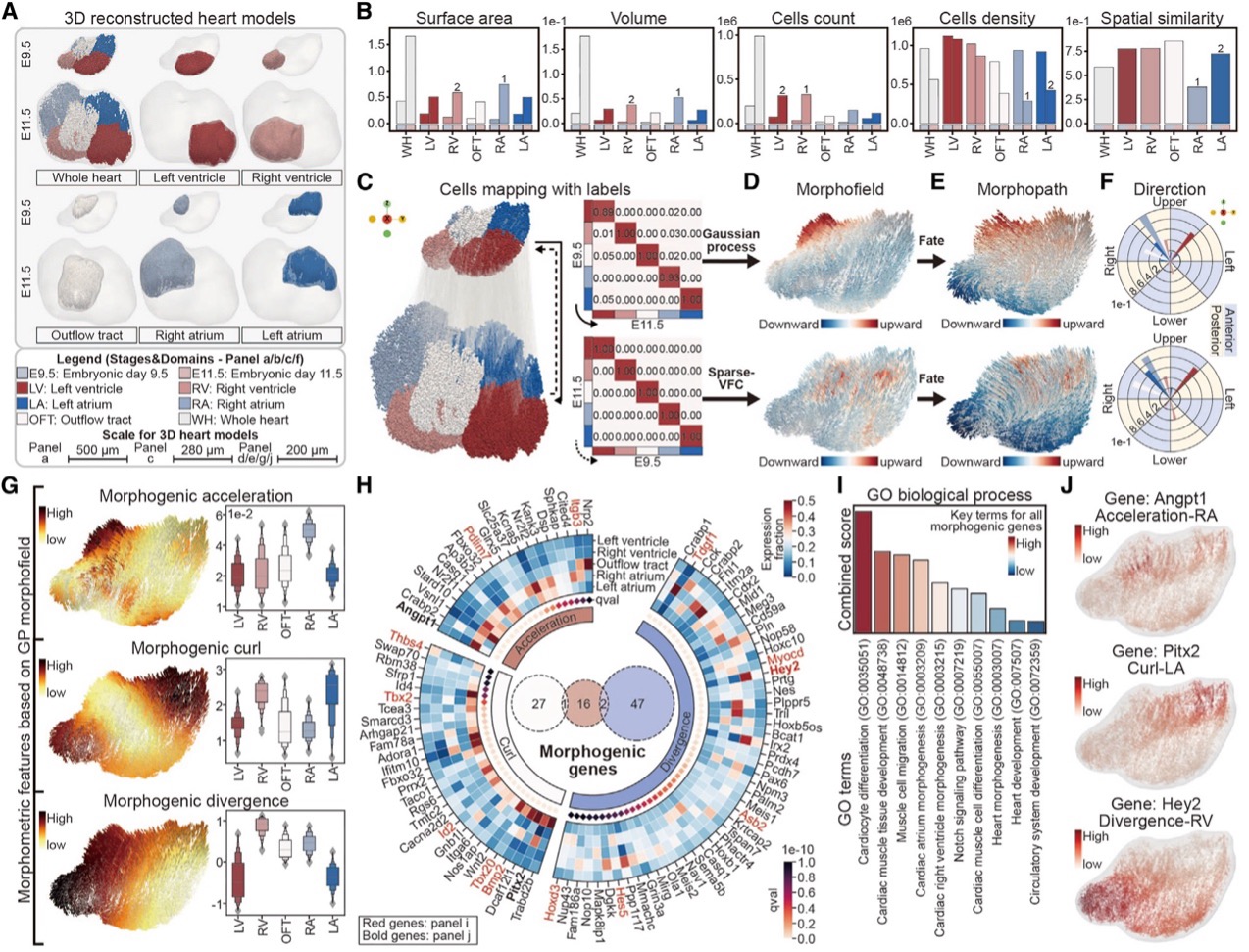 Spateo characterizes morphometric and molecular dynamics involved in the asymmetrical heart organogenesis.
Spateo characterizes morphometric and molecular dynamics involved in the asymmetrical heart organogenesis.
Expanding the Horizons of Spatial Genomics
As spatial transcriptomics technology continues to evolve, the researchers foresee numerous possibilities for applying Spateo to a wide range of biological systems, including the potential for cross-species comparisons of organ development, which could provide insights into how structures like the heart have evolved over time. The team envisions that Spateo could also be used to study complex 3D tissues involved in human diseases. As spatial technologies mature, many single-cell genomics techniques are expected to be adapted for use in spatial genomics, enabling researchers to study cells in their natural 3D environments. The Spateo framework could be instrumental in understanding the dynamic processes that govern cell behavior, helping to unlock new treatments for diseases and laying the foundation for future research in the life sciences.
Dr. Bai Yinqi, co-corresponding author of the study and a researcher at BGI-Research, emphasized that while artificial intelligence has profoundly impacted life sciences research, traditional mathematical modeling still holds unique advantages in terms of model interpretability and generalizability. He stressed that model construction should prioritize addressing biological questions, rather than simply transplanting algorithms from medical imaging fields into spatial genomics.
Dr. Xu Xun, co-corresponding author and Director of the BGI-Research, commented, "Spateo represents a major leap forward in the study of life from a spatiotemporal perspective. By drawing on theories from multiple disciplines, we have developed an algorithm that addresses challenges that experimental techniques could not solve—such as compensating for minor deformations in tissue slices, overcoming data loss caused by discontinuous sampling, and reconstructing processes like cell differentiation and migration over time. With the advancement and widespread adoption of spatiotemporal technologies, we believe this forward-looking algorithm research will lay the foundation for systematically studying life processes from a spatiotemporal dimension.”
Other senior members of the team also believe that Spateo represents a key milestone in our journey toward creating a predictive virtual embryo model capable of simulating gene or cell perturbations and predicting subsequent changes in gene expression regulation, both within cells and across spatial contexts. This model has the potential to reveal disruptions in the microenvironment, developmental trajectories, and morphological structure formation. Unlike other international ‘virtual cell’ efforts, our ‘virtual embryo’ approach is holistically defined by considering the biological system as an interconnected whole rather than focusing on individual cells, incorporating essential biological processes such as cell migration, cell-cell communication, and gene regulation across time and space. This comprehensive model holds tremendous potential for unraveling the molecular mechanisms underlying congenital diseases, offering profound implications for advancing human health.
Ethical review approval was obtained for this study. International collaborations were limited to computational modeling and data analysis, but did not involve any physical or financial resources, strictly following laws and regulations in U.S. and China. All software development in the project was carried out in a transparent manner and publicly accessible on GitHub, and the data used in this publication is now publicly accessible.
This study can be accessed here: https://www.cell.com/cell/fulltext/S0092-8674(24)01159-0
For more information about Spateo and its applications, please visit https://spateo-release.readthedocs.io/en/latest/, and the toolkit can be found here: https://github.com/aristoteleo/spateo-release. A Google Earth-like interactive Data explorer can be found at: https://viewer.spateo.aristoteleo.com/ or whose source code is in https://github.com/aristoteleo/spateo-viewer.



