March 31, Shenzhen - A recent study led by BGI-Research presents 3,324 high-quality draft genomes from isolates selected from a large-scale cultivation of bacterial isolates from fecal samples of healthy volunteers, which will help researchers better understand the process of dietary fibers metabolism and short-chain fatty acid synthesis of human gut bacteria. This study discovered valuable secondary metabolite biosynthetic gene clusters of the gut microbiota, and constructed phage–bacterium linkages in the human gut. This study was published in Nature Communications.
 The genomic landscape of reference genomes of cultivated human gut bacteria published in Nature Communications.
The genomic landscape of reference genomes of cultivated human gut bacteria published in Nature Communications.
The human body comprises human cells and symbiotic microorganisms, including bacteria, archaea, fungi, and viruses. In the human gut, the number of bacterial cells can reach 1013, which is equivalent to the total number of human cells.
In 2019, a study led by BGI-Research presented a collection of 1,520 nonredundant, high-quality draft genomes, termed Culturable Genome Reference (CGR), which was the largest collection of human gut bacterial genomes in the world at that time.
Four years later, the research team presented CGR2, a continuous work of the previous study, offering a larger bacteria bank. By integrating the data from the CGR, this project analyzed 144 fecal samples newly collected from healthy volunteers and obtained 3,324 high-quality bacteria draft genomes from more than 20,000 bacterial strains cultured under nearly 40 conditions.
The CGR2 contains 527 species-level clusters in eight phyla, and identifies 179 clusters that have never been reported previously. Compared with the CGR data, CGR2 reports a new addition of three phyla and 189 clusters.
This study also provides a comprehensive map of intestinal bacteria functions for carbohydrate metabolism. It shows that 193 strains belonging to 42 genera have complete dietary fiber (including pectin, cellulose, and inulin) metabolism and short-chain fatty acid synthesis pathways, which play an important role in human health.
Bifidobacterium, one of the most abundant probiotics, is widely harbored in the intestine of breastfed infants. The study shows that the bifidobacteria strains cultured by the research team also have a complete pathway for the metabolism of human milk oligosaccharides, which further supports the role of human milk oligosaccharides as prebiotics in regulating gut microbiota and improving human health.
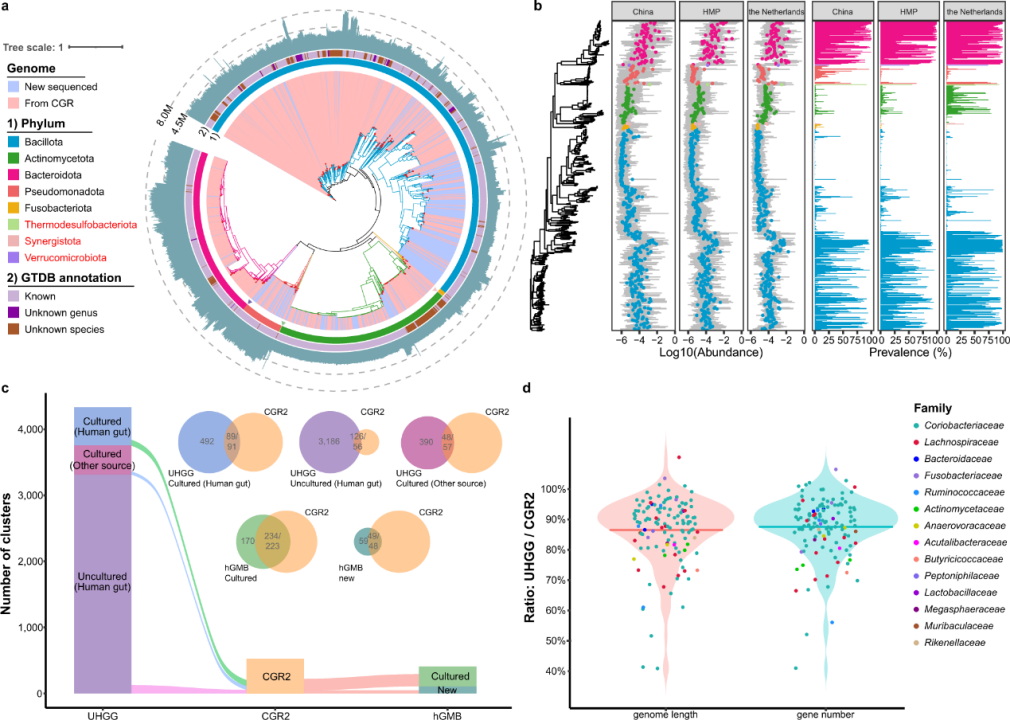 Taxonomic profile of CGR2.
Taxonomic profile of CGR2.
This study demonstrates that gut microorganisms also have a rich and diverse potential for secondary metabolite synthesis. Previously, researchers mainly explored secondary metabolites in marine or soil microbial resources. This study suggests that human gut microorganisms can serve as sources of active substances such as antimicrobial peptides and protease inhibitors, which is critical for drug research and development.
Secondary metabolites are organic compounds produced by any life form that are not directly involved in the normal growth, development, or reproduction of the organism. They are non-essential substances for bacterial growth but have a wide range of applications in medicine, agriculture, and food.
The study uncovered the phage–bacterium network in the human gut, which help to maintain the stability of the gut microbiome. Based on the CGR2 dataset, the study predicted 2,820 high-quality bacteriophage sequences, most of which had not been previously reported. Importantly, CGR2 was able to provide accurate information about the hosts for these bacteriophages.
By demonstrating the phage–bacterium linkages, the researchers revealed a complex intervention between both and found that over half of the bacteriophages were capable of infecting multiple bacterial species. Notably, the study discovered four viral clusters that can infect bacteria from different phyla, revolutionizing the current understanding of the specificity of bacteriophages.
The study found evidence that bacteriophages may mediate horizontal gene transfer events between host bacteria, which can lead to the spread of antibiotic-resistance genes and other harmful genes.
This research provides important knowledge and data for future studies on the in vitro functions of the human microbiota and its functional products, as well as the development and industrialization of probiotics. Ethical review approval was obtained for this study.
Learn more about the research: https://www.nature.com/articles/s41467-023-37396-x
Source:
[1]. Zou Y, Xue W, Luo G, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses[J]. Nature biotechnology, 2019, 37(2): 179-185.
[2]. Sender R, Fuchs S, Milo R. Revised estimates for the number of human and bacteria cells in the body[J]. PLoS biology, 2016, 14(8): e1002533.
[3]. Li S, Wang P, Yuan W, et al. Endocidal regulation of secondary metabolites in the producing organisms[J]. Scientific reports, 2016, 6(1): 29315.



