In a major scientific breakthrough, BGI-Research in collaboration with the Center for Excellence in Brain Science and Intelligence Technology at the Chinese Academy of Sciences, Lingang Laboratory, and Tencent AI Lab has successfully constructed a high-resolution spatial transcriptomic atlas of the adult mouse brain using cutting-edge whole-genome spatial transcriptomic techniques with single-cell resolution. This remarkable achievement, recently published in the journal Neuron, provides an entirely new spatial perspective for understanding the complex functions of the mammalian brain by mapping nearly 30,000 genes and more than four million cells into 308 distinct cellular clusters.
 The study “Single-cell spatial transcriptomic atlas of the whole mouse brain” was published in Neuron.
The study “Single-cell spatial transcriptomic atlas of the whole mouse brain” was published in Neuron.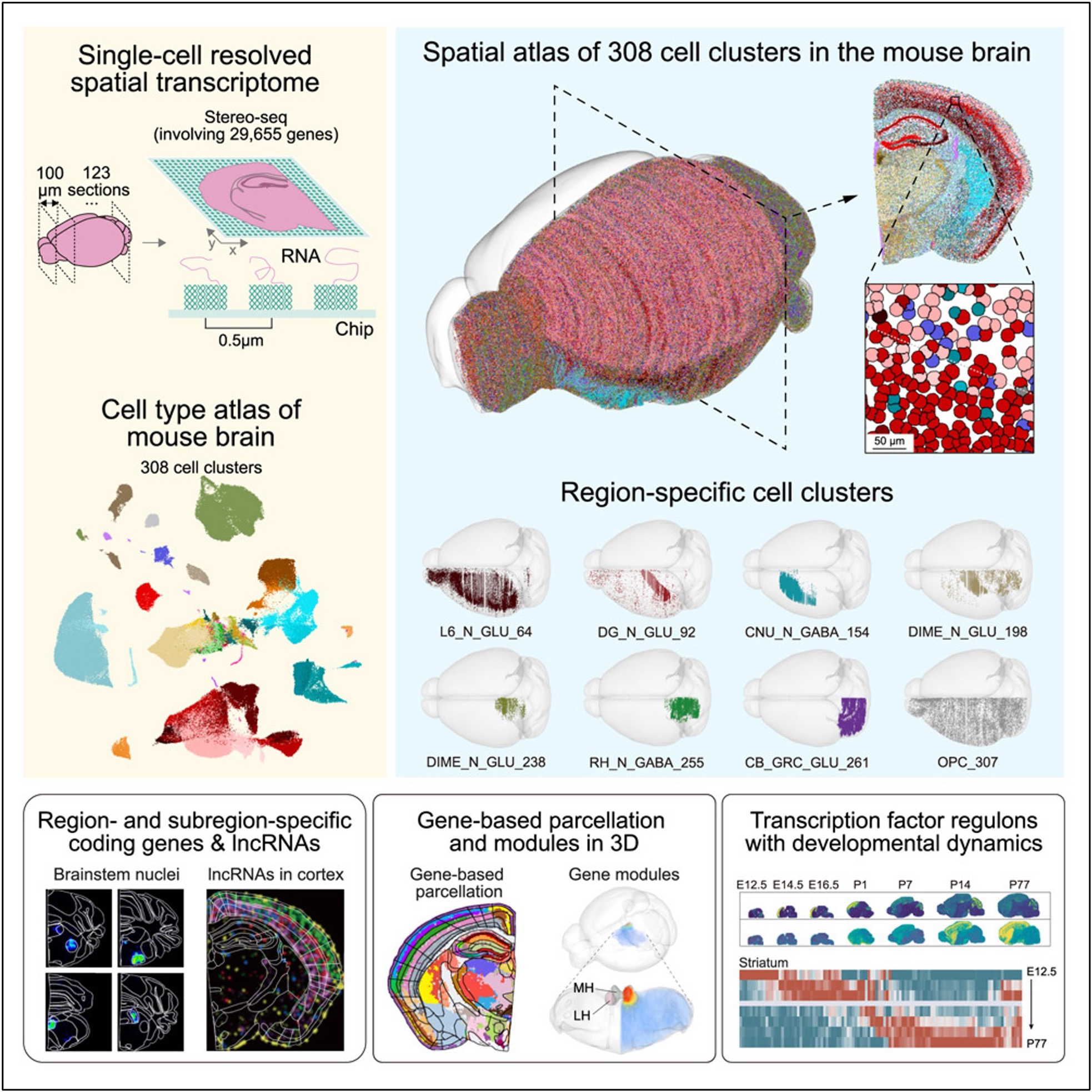 Graphical abstract of this study.
Graphical abstract of this study.
The study stands out by revealing the regional specificity and spatiotemporal dynamics of non-coding RNAs, particularly long non-coding RNAs (lncRNAs), within the adult and developing mouse brain. By integrating spatial expression data for lncRNAs for the first time, the research uncovers how these molecules exhibit distinct regional enrichment and dynamic changes throughout development, offering fresh clues into their roles in brain function and in the etiology of neurological diseases. The ability to capture such detailed non-coding transcriptomic profiles represents a significant advance over traditional methods that primarily focused on protein-coding genes.
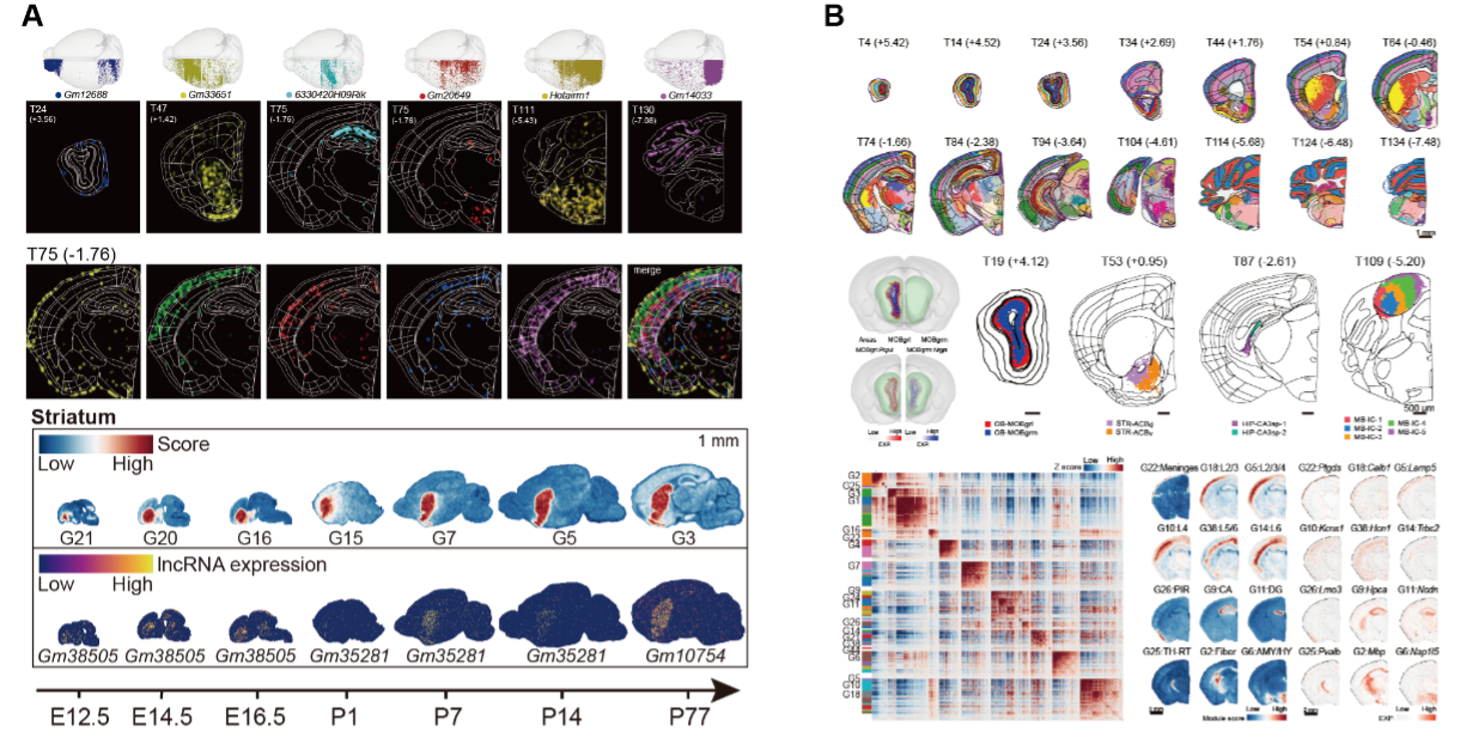 Certain non-coding RNAs are enriched in specific brain regions and change during development, suggesting they may shape brain function.
Certain non-coding RNAs are enriched in specific brain regions and change during development, suggesting they may shape brain function.
In addition to the novel insights into non-coding RNAs, the research demonstrates how region-specific gene expression can be leveraged to optimize brain parcellation. Traditionally, brain regions have been defined solely by cytoarchitecture and function, yet this study employs whole-genome spatial transcriptomics to refine these boundaries by incorporating transcriptomic signatures. The use of advanced spatial clustering methods not only validates the transcriptomic basis for brain region segmentation but also reveals gene modules that are uniquely associated with specific areas. These findings pave the way for new methodologies to study the complex molecular architecture of brains in other species as well.
Another key highlight of this work is the revelation of critical regulatory factors driving brain development. By analyzing data from seven distinct developmental stages ranging from the embryonic period to adulthood, the research team identified 411 transcription factors with significant spatiotemporal variations. These transcription factor regulons display finely tuned expression patterns along developmental axes, such as the rostral-caudal and dorsal-ventral gradients, which are closely linked to the formation of intricate cortical substructures. This comprehensive temporal map provides unprecedented insights into the regulatory networks that shape brain development and function.
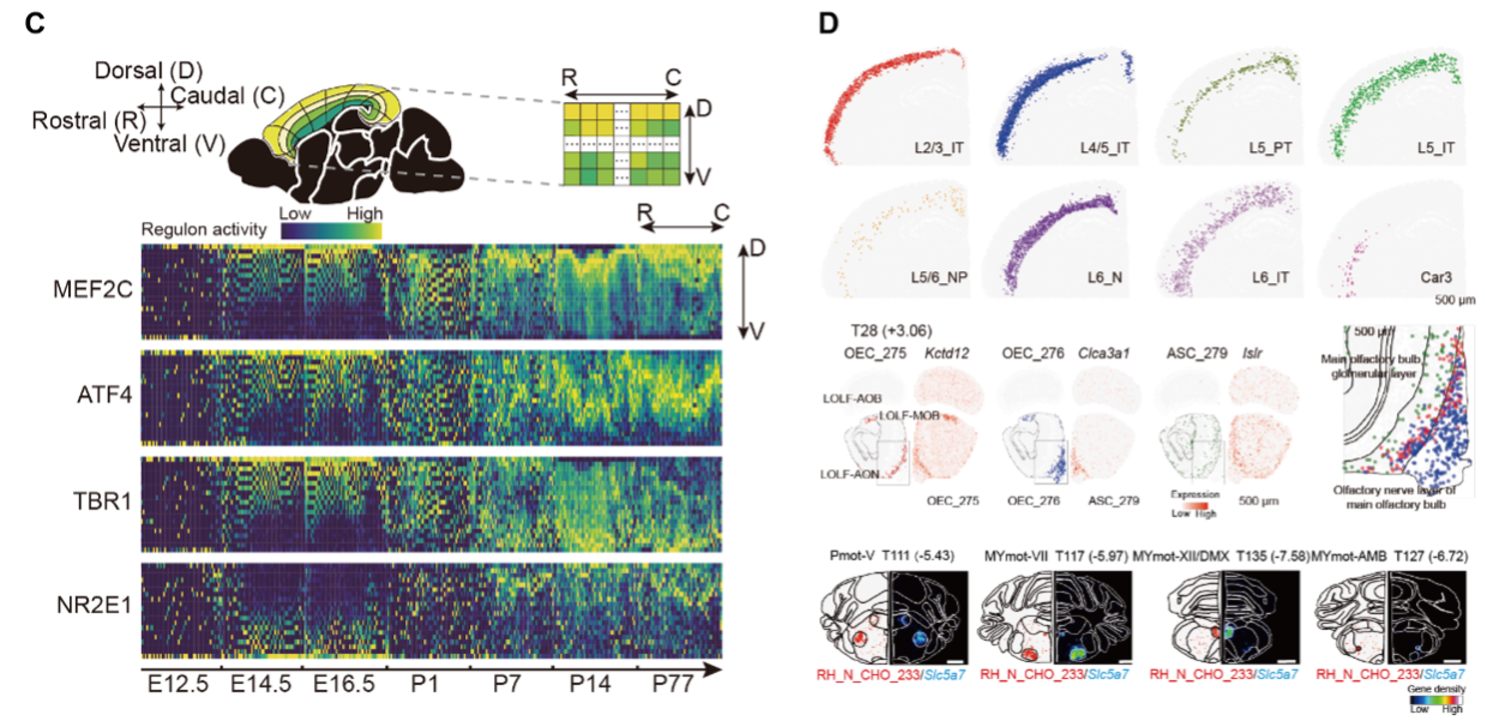 As the mouse cortex matures, the activity of key transcription factors shifts, revealing how brain layers form over time.
As the mouse cortex matures, the activity of key transcription factors shifts, revealing how brain layers form over time.
The atlas also sheds light on the spatial heterogeneity of cell types within the mouse brain, including the discovery of diverse neuronal subtypes and regionally enriched non-neuronal cells. For instance, the study highlights variations in motor neuron subtypes within different brainstem motor nuclei, offering crucial clues to the molecular mechanisms underlying motor control. Such detailed cellular mapping not only deepens our understanding of the functional organization of the brain but also holds promise for advancing research into neurological disorders.
This research highlights the potential of integrating multi-dimensional transcriptomic data with advanced spatial analysis to better understand the complexity of the brain, providing a valuable tool for both foundational studies and therapeutic innovation. Emphasizing the importance of collaborative efforts, the study showcases meaningful advancements in brain research, with contributions from BGI playing a critical role.
Ethical review approval was obtained for this study.
This research can be accessed here: https://www.cell.com/neuron/abstract/S0896-6273(25)00133-3



