On April 6, in a new study published in Molecular Plant, researchers from BGI-Research, Huazhong Agricultural University, and their collaborators have pinpointed key genes linked to drought resistance in rice, with HMGB1 emerging as a critical transcriptional regulator of root development. Utilizing BGI’s advanced spatial transcriptomics technology, Stereo-seq, the team mapped 376 genes involved in root development that likely contribute to stronger roots of upland rice that enhance its survival in dryland conditions. These findings illuminate the genetic mechanisms driving drought adaptation and suggest a potential pathway to convert irrigated rice into drought-tolerant varieties.
 The study “Comparative spatial transcriptomics reveals root dryland adaptation mechanism in rice and HMGB1 as a key regulator” was published in Molecular Plant.
The study “Comparative spatial transcriptomics reveals root dryland adaptation mechanism in rice and HMGB1 as a key regulator” was published in Molecular Plant.
Drought currently affects over 40% of the world’s land area, with regions such as sub-Saharan Africa and South Asia facing severe agricultural challenges. The United Nations predicts that by 2030, 700 million people could face displacement due to water scarcity, exacerbated by climate change and prolonged dry spells that devastate global agriculture. As a staple food for more than half the world’s population, rice faces an urgent need for resilient varieties.
Unlike widely grown irrigated rice, upland rice - a unique geographical variant cultivated in about one-sixth of rice-producing areas - thrives in dryland and nutrient-poor soils. It exhibits fewer crown roots, longer root systems, and thicker root structures. These adaptations, driven by its robust root architecture, enable it to access deeper soil moisture reserves. Until now, however, the molecular underpinnings of these traits have remained largely unclear.
 Root systems comparison of irrigated rice and upland rice varieties.
Root systems comparison of irrigated rice and upland rice varieties.
The research team examined 16 rice cultivars, including four upland and four irrigated japonica varieties, as well as four upland and four irrigated indica varieties. Japonica and indica are the two primary domesticated types of Asian rice. Upland japonica consistently showed fewer crown roots, greater root length, and thicker roots compared to its irrigated counterparts, while these traits were less pronounced in the indica group. This suggests that upland japonica and indica rice rely on distinct strategies to cope with water scarcity.
The application of Stereo-seq technology proved instrumental in this research, enabling the team to create detailed molecular maps of rice root development. By leveraging Stereo-seq, researchers provided a clear spatial structure of the coleoptilar node, capturing the transcriptional signatures of thousands of genes. The research team charted the anatomical structure of the transition zone between stem and root for the first time, offering an unprecedented view of this critical region. This approach identified 376 genes linked to root development that are also differentially expressed between irrigated rice and upland rice, with HMGB1 standing out as a key regulator of root growth and drought resilience.
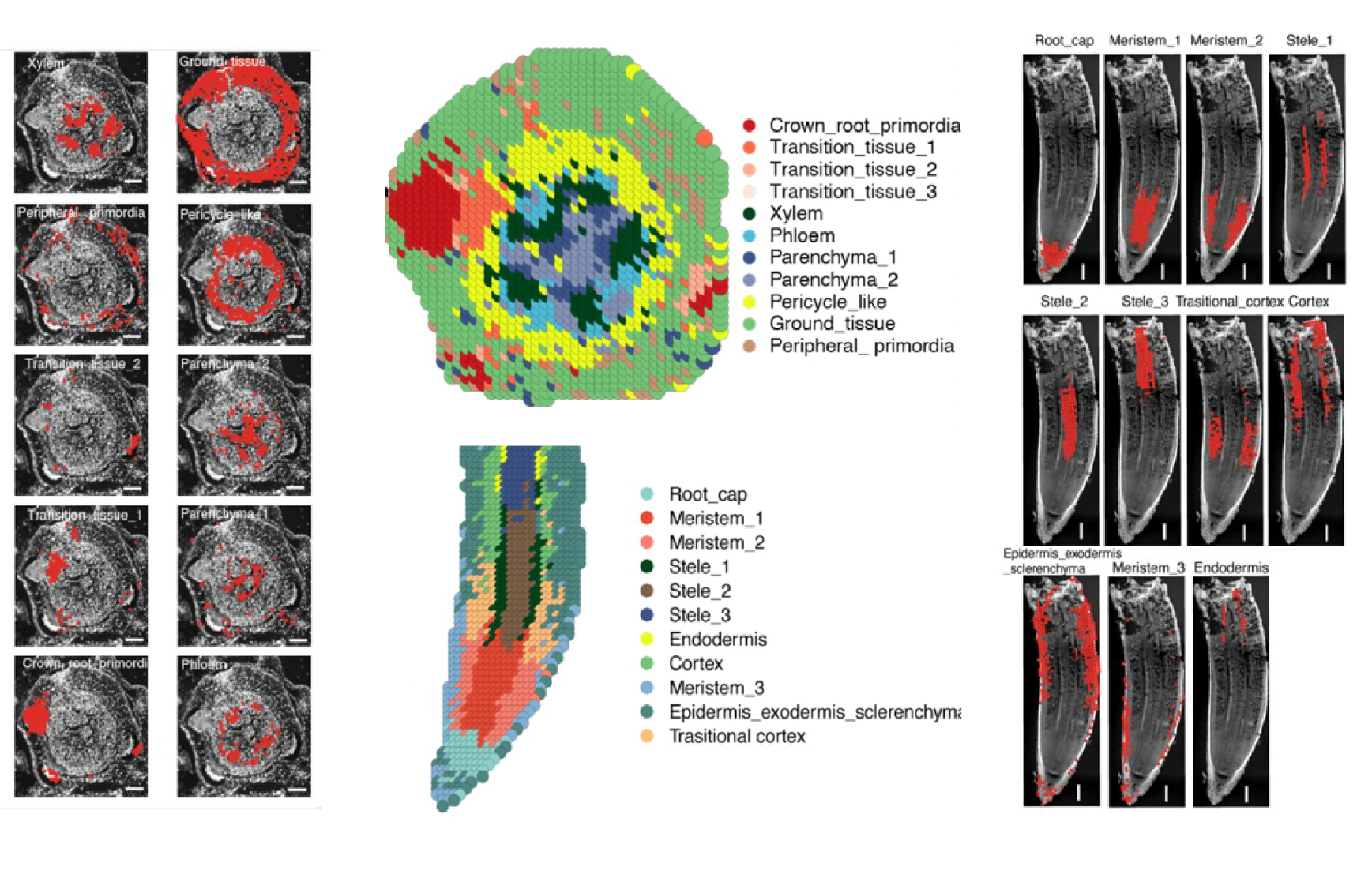 Spatial transcriptomic atlas of coleoptilar node and root tip.
Spatial transcriptomic atlas of coleoptilar node and root tip.
In upland rice, lower HMGB1 expression correlated with longer roots, larger meristem zones, and thicker cortical cell layers - features that improve water access in deeper soil layers. The spatially resolved data clarified the genetic foundation of upland rice’s specialized root architecture and provided a transcriptomic timeline of crown root initiation, a process vital for dryland adaptation.
To confirm HMGB1’s role, the team removed this gene in irrigated rice. The resulting plants developed traits resembling upland rice, including longer roots and enhanced drought tolerance. This finding suggests that modulating HMGB1 expression could transform irrigated rice into drought-resilient varieties capable of thriving in water-scarce environments.
 Comparison of HMGB1 removed (hmgb1) and HMGB1 overexpressing (OEHMGB1) phenotypes.
Comparison of HMGB1 removed (hmgb1) and HMGB1 overexpressing (OEHMGB1) phenotypes.
The discovery of HMGB1 and other drought-resistance genes provides a valuable genetic toolkit for breeding rice varieties that can endure harsh conditions. By applying these insights, scientists and breeders can develop cultivars that sustain yields despite dwindling water resources, bolstering food security for millions of farmers and consumers worldwide.
This study builds on decades of progress in rice genomics. In 2002, BGI and its partners published a draft sequence of the indica rice genome in Science, a milestone in rice research. By 2005, BGI-led scientists published the fine map of the rice genome in PLoS Biology. These foundational efforts supplied the genomic tools and frameworks that continue to propel discoveries in rice biology, including this landmark research on drought resistance.
This research not only deepens our understanding of how upland rice adapts to dryland environments but also lays a practical foundation for enhancing crop resilience. As environmental pressures intensify, the insights gained from this study could ensure that rice - a cornerstone of global diets - remains a reliable and sustainable food source in an increasingly unpredictable climate.
This research can be assessed here: https://www.sciencedirect.com/science/article/pii/S1674205225001108



