April 11, Shenzhen - A team of scientists from BGI-Research, Southern University of Science and Technology and other institutes has unveiled the most detailed single-cell map of a plant ever created, offering new insight into how plants grow, aging, and efficiently recycle their nutrients. Using BGI’s innovative single-cell sequencing technologies, the study reveals that leaf aging - known as senescence - is not simply a process of decay but a carefully coordinated event where older leaves give up their nutrients to support new growth. Published in Cell, this breakthrough provides a new understanding of plant physiology at a level of detail that was previously impossible.
 The study, titled “An Arabidopsis single-nucleus atlas decodes leaf senescence and nutrient allocation,” was published in Cell.
The study, titled “An Arabidopsis single-nucleus atlas decodes leaf senescence and nutrient allocation,” was published in Cell.
Over the past six years, many single-cell atlases have been developed for specific organs, mostly using a method called protoplasting, which involves removing the cell wall to study the interior. However, this method struggles with older leaves and reproductive organs like flowers and siliques, where cells are tougher and more difficult to isolate. To overcome this, the team used single-nucleus sequencing, which bypasses the need to extract whole cells and instead focuses on the nucleus, allowing for better preservation of delicate tissues.
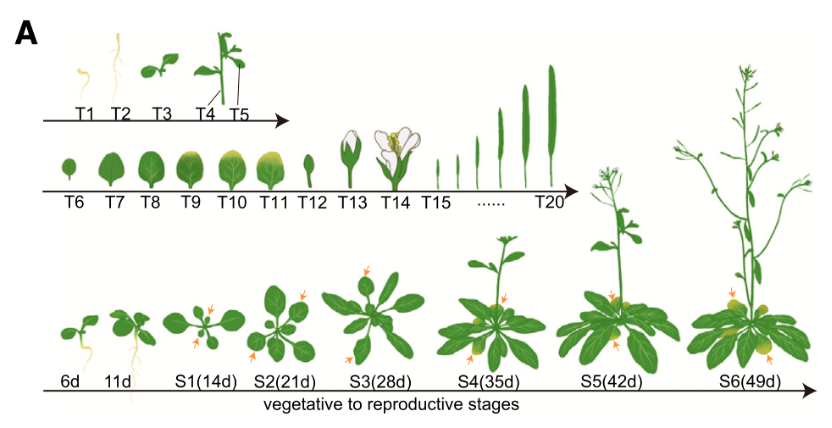 A total of 20 tissues (T1–T20) were collected from vegetative growth to reproductive growth, with six stages of the second pair of true leaves are indicated by arrows.
A total of 20 tissues (T1–T20) were collected from vegetative growth to reproductive growth, with six stages of the second pair of true leaves are indicated by arrows.
By analyzing 913,769 individual plant cell nuclei from 20 distinct tissues, researchers constructed a full life-cycle map of Arabidopsis thaliana, a small flowering plant commonly used in scientific research. This map spans from roots and stems to leaves, flowers, and siliques (seed pods), capturing how different cell types behave across time and space.
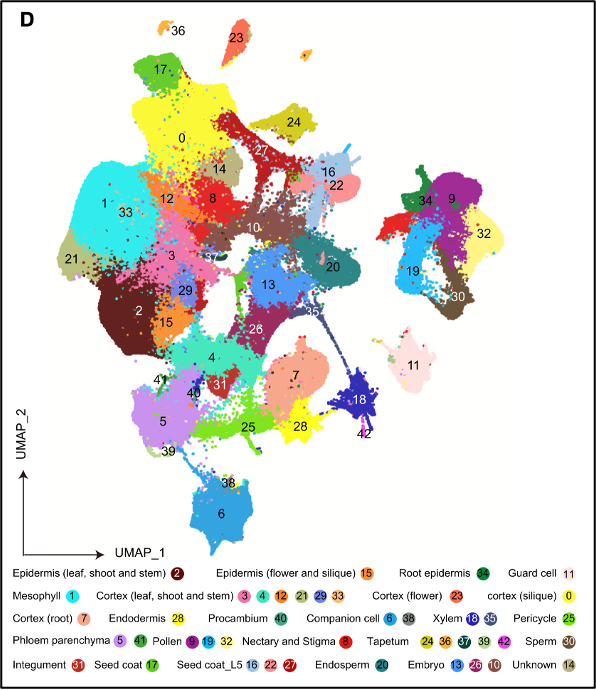 The single-nucleus atlas of Arabidopsis colored by major cell types.
The single-nucleus atlas of Arabidopsis colored by major cell types.
The researchers identified 38 distinct cell types from their analysis, including several that are shared across different organs and others that are organ-specific. This comprehensive map enables scientists to better understand how different parts of the plant coordinate their functions - especially during aging, which is a critical phase in the plant’s life cycle.
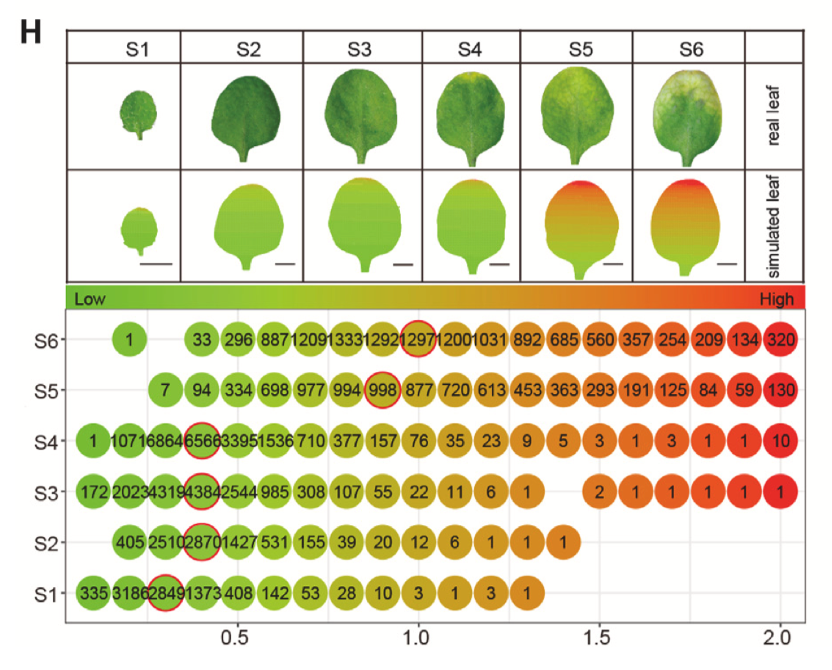 Simulated leaves and cell alignment based on the SAG-index values of all the leaf cells along the six sampling stages. Numbers in dots indicate the counts of cells with indicated SAG-index value. Red circles mark the cell population with median SAG-index value of each sampling stage.
Simulated leaves and cell alignment based on the SAG-index values of all the leaf cells along the six sampling stages. Numbers in dots indicate the counts of cells with indicated SAG-index value. Red circles mark the cell population with median SAG-index value of each sampling stage.
One of the most exciting results from the study is the creation of two new molecular tools: the Senescence-Associated Gene (SAG) Index and the Youth-Associated Gene (YAG) Index. These indexes measure how “old” or “young” a plant cell is based on its gene activity. The YAG Index peaked during early leaf development and gradually declined as the leaf matured. Meanwhile, the SAG Index remained low until the leaf began to show visible signs of aging - such as yellowing at the tips - after which it rose sharply.
Surprisingly, even in leaves that appeared completely green, many cells already showed high SAG Index values, hinting that the aging process had begun much earlier than visible symptoms appeared. The presence of youth cells in visibly old leaves also suggests that leaf aging is not uniform, but rather a mosaic of aging and non-aging cells. This finding reshapes how scientists understand the timing and regulation of plant senescence.
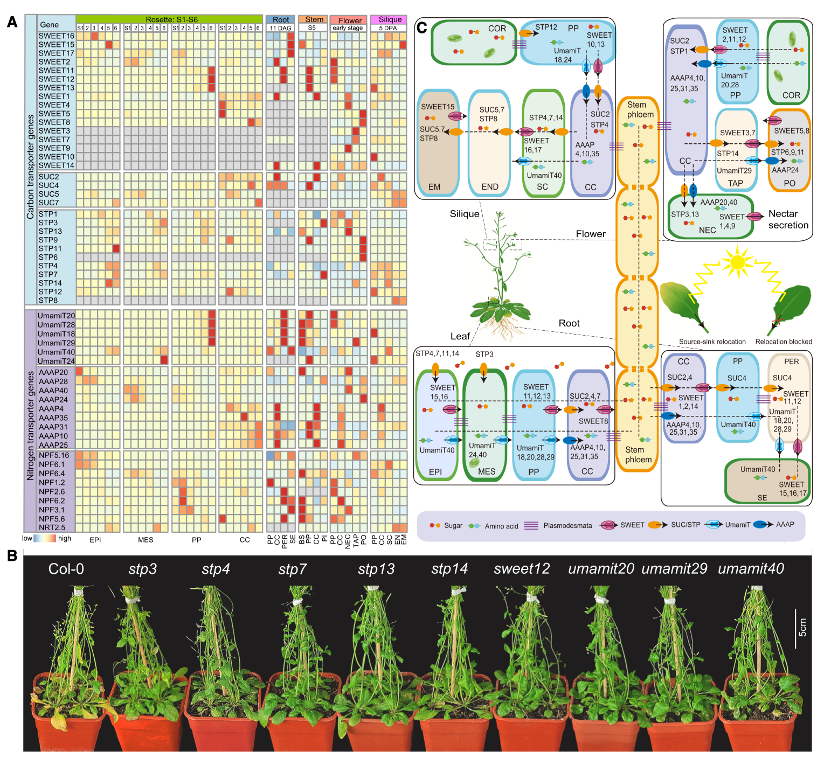 Dynamic allocation of carbon and nitrogen in leaves and reproductive organs.
Dynamic allocation of carbon and nitrogen in leaves and reproductive organs.
The study also delves into the complex journey of nutrients during leaf aging. As leaves grow old, they do not waste their accumulated sugars and proteins. Instead, they transport these nutrients to parts of the plant that are still growing - such as flowers, roots, and developing seeds. This recycling process is vital to the plant’s survival and reproduction.
The team discovered that specific transport proteins are responsible for this nutrient movement. Sugar transporter proteins known as SWEET and SUCs/STP were found to be active in certain cells in aging leaves, moving sugars from leaves to flowers and siliques, where they fuel the growth of new seeds. Similarly, proteins from the UmamiT and AAAP family were found to move nitrogen-rich amino acids from aging leaves to other organs. These proteins were especially active in specialized vein cells, acting like molecular highways for nutrient transfer.
In root and stem tissues, related transporters performed similar roles, highlighting the importance of cell-type-specific nutrient movement throughout the entire plant. The study suggests that a cell’s location and identity determine how it participates in the plant’s overall nutrient economy, with different organs contributing in unique ways to the recycling process.
For the scientific community, this publication offers a valuable dataset that can be explored for years to come, deepening our understanding of plant development and cellular function. At the same time, for the general public and agricultural sector, it offers new hope - this cellular-level insight into nutrient allocation opens up exciting possibilities for more sustainable and productive farming. By regulating the activity of key transporters or adjusting the timing of leaf senescence, scientists may breed crops that use nutrients more efficiently, require less fertilizer, and yield more food. Fine-tuning the aging process could also extend the productive life of leaves, supporting better nutrient flow to seeds and fruit without sacrificing crop quality.
This research can be accessed here: https://doi.org/10.1016/j.cell.2025.03.024



