Breaking new ground in spatial transcriptomics, BGI has unveiled Stereopy, an innovative analytical framework that paves the way for a new era in deciphering cellular complexity across multiple samples. Published in Nature Communications, the study details how Stereopy not only resolves the challenges of multi-sample spatial transcriptomics but also provides a robust and standardized framework that can reveal subtle biological mechanisms underpinning complex systems.
 The study titled “Stereopy: Modeling Comparative and Spatiotemporal Cellular Heterogeneity via Multi-Sample Spatial Transcriptomics” was published in Nature Communications, showcasing a breakthrough study in multi-sample spatial transcriptomics that provides new insights into cellular heterogeneity.
The study titled “Stereopy: Modeling Comparative and Spatiotemporal Cellular Heterogeneity via Multi-Sample Spatial Transcriptomics” was published in Nature Communications, showcasing a breakthrough study in multi-sample spatial transcriptomics that provides new insights into cellular heterogeneity.
Spatial transcriptomics has long promised to transform our understanding of tissue development and disease mechanisms by simultaneously mapping gene expression and cellular location, yet most existing tools were designed for single-sample analysis and therefore fell short when it came to comparative studies, time-series investigations, and three-dimensional tissue reconstructions. Recognizing this critical gap, BGI’s research team has developed Stereopy to offer a comprehensive solution that integrates advanced data management, analysis, and visualization capabilities into one unified platform.
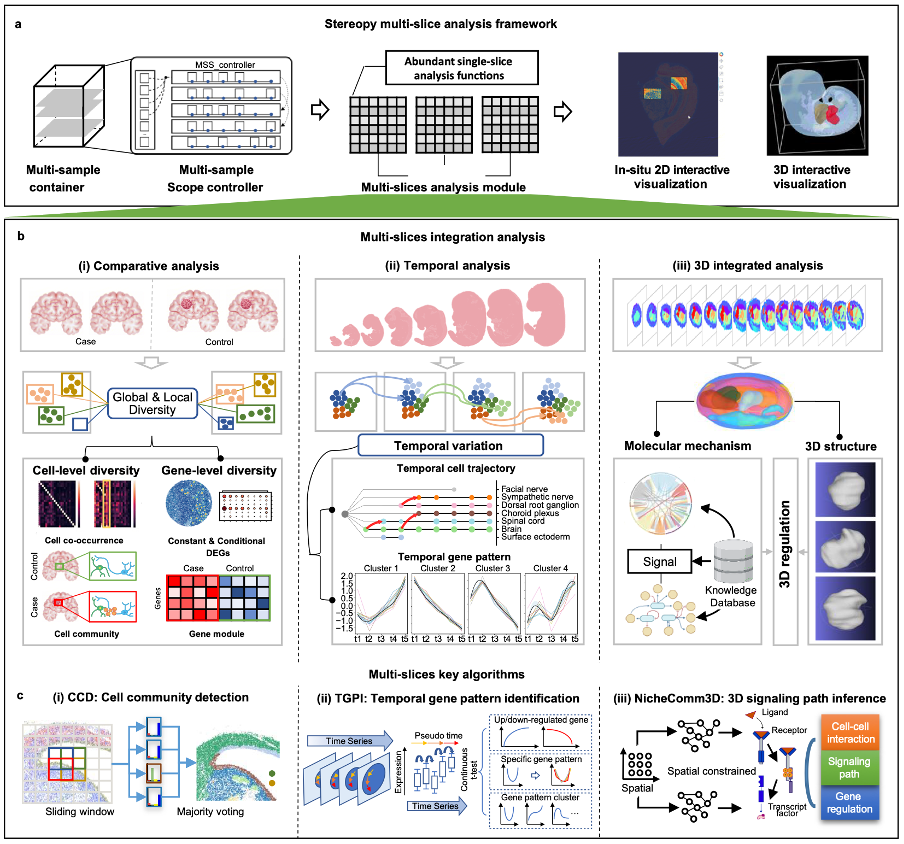 The overall framework of Stereopy. It provides a comprehensive solution for multi-sample analysis including a data container and analytical framework, spatial omics analysis modules, and interactive visualization.
The overall framework of Stereopy. It provides a comprehensive solution for multi-sample analysis including a data container and analytical framework, spatial omics analysis modules, and interactive visualization.
At the heart of Stereopy lies a trio of core components that work in concert to transform the way researchers approach spatial omics data. The MsData container enables seamless integration and flexible management of vast datasets while preserving the unique characteristics of individual samples. Complementing this is the MSS controller, an intelligent system that tracks the interdependencies of analysis outcomes and delivers interactive visualizations with a single click, thereby simplifying complex data workflows. Further enhancing its versatility, the multi-sample transformation module MS-ST allows researchers to either integrate data for a holistic view or dissect it to examine localized features, ensuring that both global and regional variations are meticulously captured.
This integrated framework has been rigorously benchmarked and has demonstrated superior efficiency and accuracy compared to existing analytical tools, thereby underscoring its potential to drive breakthrough discoveries in areas such as brain development and tumor microenvironments.
 Multi-sample omics analysis framework of Stereopy featuring the MsData container, MSS control hub, and MS-ST analysis transformer, offering diverse approaches for spatial omics research.
Multi-sample omics analysis framework of Stereopy featuring the MsData container, MSS control hub, and MS-ST analysis transformer, offering diverse approaches for spatial omics research.
Stereopy’s transformative impact is further driven by three innovative algorithms specifically designed to address distinct analytical challenges. The Stereopy-CCD algorithm excels in comparative analysis by precisely delineating cell community differences between normal and disease samples, thereby uncovering tissue-specific biomarkers and functional mechanisms. Meanwhile, the Stereopy-TGPI algorithm brings a new dimension to time-series analysis by jointly assessing spatial distribution and temporal changes in gene expression, enabling the construction of dynamic regulatory networks that illuminate the processes of development and disease progression. The Stereopy-NicheReg3D algorithm represents another leap forward by reconstructing three-dimensional cellular microenvironments at single-cell resolution and inferring intricate ligand-receptor interactions and downstream regulatory pathways. Together, these algorithms not only offer unparalleled insights into the spatiotemporal dynamics of cellular systems but also set new standards for accuracy and comprehensiveness in the field.
"Stereopy is not only an innovation in technical tools but also a revolution in the scientific research paradigm. We hope this achievement will provide strong support to the global life sciences community in jointly exploring the mysteries of life systems," said Dr. Shuangsang Fang, first author of the study and Program Scientist at BGI-Research.
 Stereopy has been integrated into BGI’s DCS Cloud platform and is readily accessible to researchers.
Stereopy has been integrated into BGI’s DCS Cloud platform and is readily accessible to researchers.
Demonstrating both its scientific rigor and practical utility, Stereopy has already been embraced by the global research community with over 48,000 downloads by March 2025, and is serving thousands of projects. Integrated into BGI’s DCS Cloud platform, Stereopy is readily accessible to researchers seeking to harness the power of spatial and single-cell transcriptomics for a wide range of applications from precision medicine to drug development.
Stereopy can be accessed at https://github.com/STOmics/Stereopy. Its documentation and extensive tutorials are available at https://stereopy.readthedocs.io/en/latest.
This research paper can be accessed here: https://www.nature.com/articles/s41467-025-58079-9



