For decades, liquid biopsy tests have analyzed the size and positioning of double-stranded cell free DNA (cfDNA) fragments in blood, but they ignore the single-stranded fragments that escape conventional detection. In a study published in Clinical Chemistry, scientists at BGI-Research report that cfDNA carries an overlooked layer of information: which strand of the fragmented DNA was released from the original cellular double-stranded DNA.
 The online publication of the research article entitled "Cell Free DNA Comprises the Strand Specific Characteristic Associated with Transcrip-tion and Methylation." in Clinical Chemistry
The online publication of the research article entitled "Cell Free DNA Comprises the Strand Specific Characteristic Associated with Transcrip-tion and Methylation." in Clinical Chemistry
By coupling a single-stranded library preparation workflow with the high accuracy DNBSEQ sequencing platform, the team captured both single- and double-stranded cfDNA from just 0.2 mL of plasma and, for the first time, kept track of which strand each fragment came from.
When they analyzed whole-genome data from 34 healthy volunteers, they discovered that certain chromosomal regions show a pronounced "strand-specific bias" where fragments from one strand vastly outnumber those from the other. The hot spots cluster in promoters, CpG islands and ac-tive genes, and remarkably, the enrichment score for these regions tracks chromosome gene density with a Spearman correlation of 0.91, under-scoring the tight link between this newly revealed signal and gene activity.
This discovery rests on the strand-by-strand maps generated with BGI's single-stranded library preparation technology. These maps highlighted that genomic elements including promoters, exons, and CpG islands were enriched in strand-specific bias windows, while regions with higher CpG densities and lower methylation levels were more likely to generate the bias.
The technology also revealed signatures in cell-free mitochondrial DNA, where the heavy strand exhibits higher read depth outside the noncoding region, while the light strand takes over exactly where transcription and replication initiate, painting a molecular picture of the organelle's asym-metric replication cycle.
Beyond nuclear and mitochondrial patterns, the method captured distinct strand-specific signatures between males and females, identifying sex differential genes enriched in pathways associated with autoimmune thyroid diseases, COVID-19, and other biologically relevant processes.
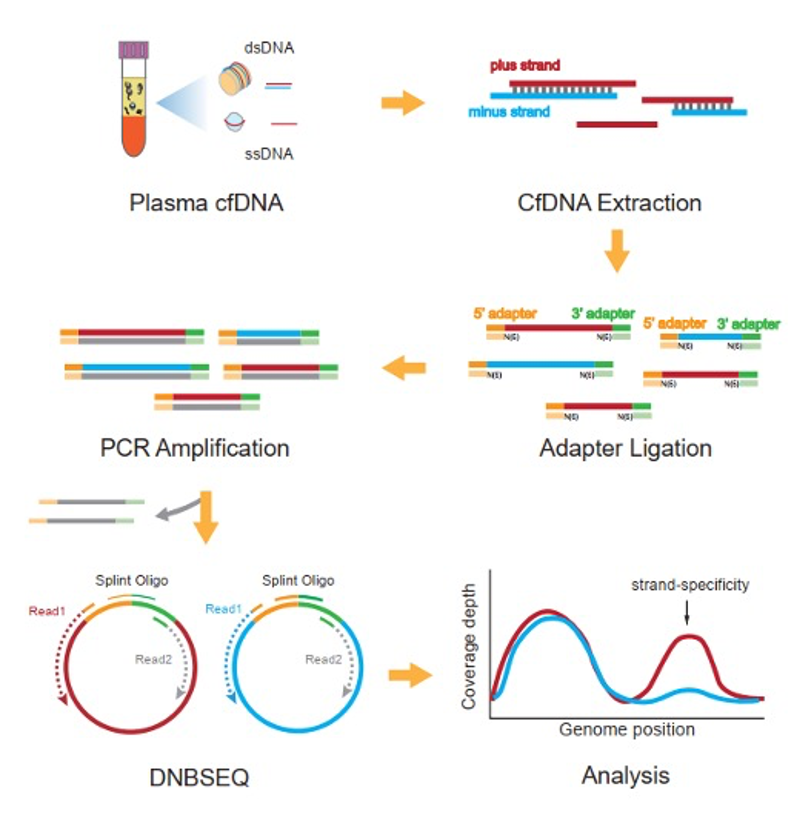 Schematic showing how BGI's single-stranded library preparation, com-bined with DNBSEQ sequencing, denatures each cfDNA fragment, ligates strand-specific adapters and preserves the directionality information that is lost in conventional double-stranded workflows.
Schematic showing how BGI's single-stranded library preparation, com-bined with DNBSEQ sequencing, denatures each cfDNA fragment, ligates strand-specific adapters and preserves the directionality information that is lost in conventional double-stranded workflows.
Extending the concept from physiology to pathology, the researchers re-analyzed an independent, low coverage dataset comprising 18 noncancer controls and 14 late-stage lung cancer patients. Even at roughly 1.2× genomic depth, a multivariate combination of strand-specific bias features separated the two groups clearly, and several promoter regions including HAR1B, CFAP410 and SNX16 showed conspicuous strand bias only in cancer plasma.
The single-stranded protocol can generate deep cfDNA whole genome sequence (30×), deep whole genome bisulfite sequence (30×) or a wide-spectrum cfRNA profile, all from a single tube of blood, opening a path toward multi omics diagnostics that could detect cancer earlier while monitoring treatment response and tracking other diseases.
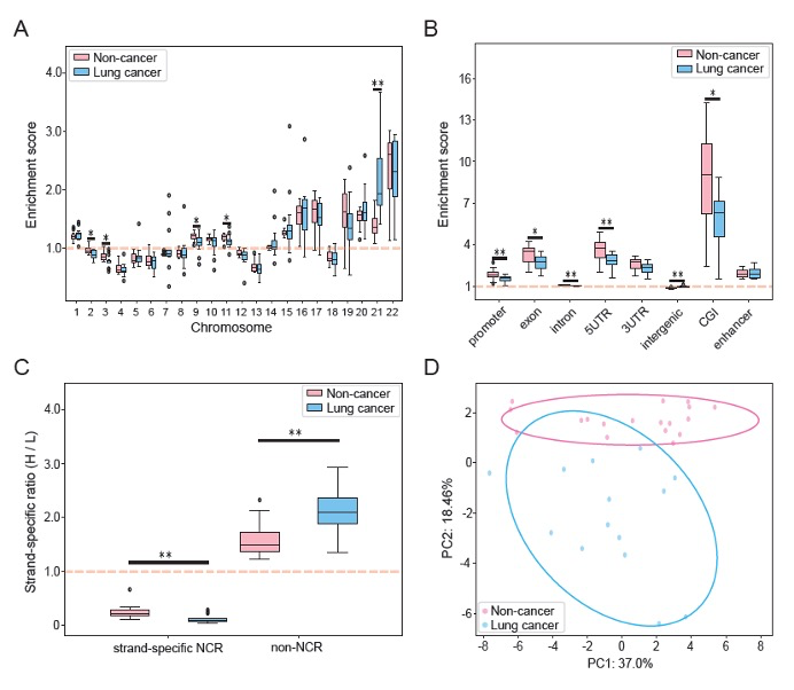 Principal component analysis reproduced from the study: a composite of strand-specific features across autosomes, genomic elements and mitochondrial DNA clearly separates late-stage lung cancer patients from noncancer controls, demonstrating the biomarker potential of the new signal even at low sequencing depth.
Principal component analysis reproduced from the study: a composite of strand-specific features across autosomes, genomic elements and mitochondrial DNA clearly separates late-stage lung cancer patients from noncancer controls, demonstrating the biomarker potential of the new signal even at low sequencing depth.
The clinical implications stretch far beyond cancer detection. Strand specificity adds a wholly new dimension to liquid biopsy panels, comple-menting existing fragment length, nucleosome footprints and methylation fingerprints. For oncologists seeking earlier detection, clinicians monitoring autoimmune diseases, or researchers tracking transplant rejection, the prospect of extracting additional diagnostic information from the same blood draw represents a significant advance in precision medicine.
This breakthrough in strand-specific fragmentomics opens new possibilities for enhancing early cancer detection, advancing prenatal testing, and improving diagnostic precision across multiple clinical applications where extracting additional molecular information from blood samples could transform patient care.
"Strand specificity is the missing compass in fragmentomics," said Dr. Yan Zhang, co-corresponding author and associated researcher at BGI-Research. "By reading not only the letters but the strand-specificity of each fragment, we can infer gene expression, DNA methylation and even mitochondrial replication activity, all without an invasive biopsy."
The study's comprehensive genomic datasets are publicly available to support continued research in liquid biopsy applications. All volunteers provided written informed consent before sampling. This research can be accessed here: https://doi.org/10.1093/clinchem/hvaf070



