In the world of genetics, it is crucial to understand where genes are active within tissues. This need has given rise to a technology known as spatially resolved transcriptomics (SRT), which merges gene expression data with precise cell locations within tissues. However, a persistent challenge in this field is the noise - unwanted random variations in the data that can obscure real results. To address this, researchers from BGI-Research developed SpotGF, an algorithm for denoising SRT data using optimal transport-based gene filtering. This study was published as the cover article in Cell Systems on October 16, 2024.
 The research “SpotGF: Denoising Spatially Resolved Transcriptomics Data Using an Optimal Transport-Based Gene Filtering Algorithm” was published as the cover article in Cell Systems.
The research “SpotGF: Denoising Spatially Resolved Transcriptomics Data Using an Optimal Transport-Based Gene Filtering Algorithm” was published as the cover article in Cell Systems.
When studying how genes are expressed in different parts of a tissue, scientists rely on SRT to pinpoint exactly where specific gene activities occur. However, the process isn’t flawless. Technical issues like cell damage during sample preparation or exposure to various chemicals can introduce noise, misleading researchers by suggesting gene activity where there might be none. This noise can severely affect crucial tasks like identifying what types of cells are present and understanding how they interact, potentially leading to incorrect scientific conclusions.
SpotGF stands out by tackling the noise directly through optimal transport-based gene filtering. This approach assesses how much gene expression data has potentially spread from its original location and filters out these misleading signals without altering the raw data. This ensures that only the in-suit gene expression signals are considered, which dramatically improves the accuracy of data analysis.
The brilliance of SpotGF lies in its ability to differentiate between noise and valuable data. Traditional methods often adjust the data to compensate for noise, which can inadvertently introduce errors, known as false positives. SpotGF avoids this pitfall by maintaining the integrity of the original data, only removing what is genuinely noise.
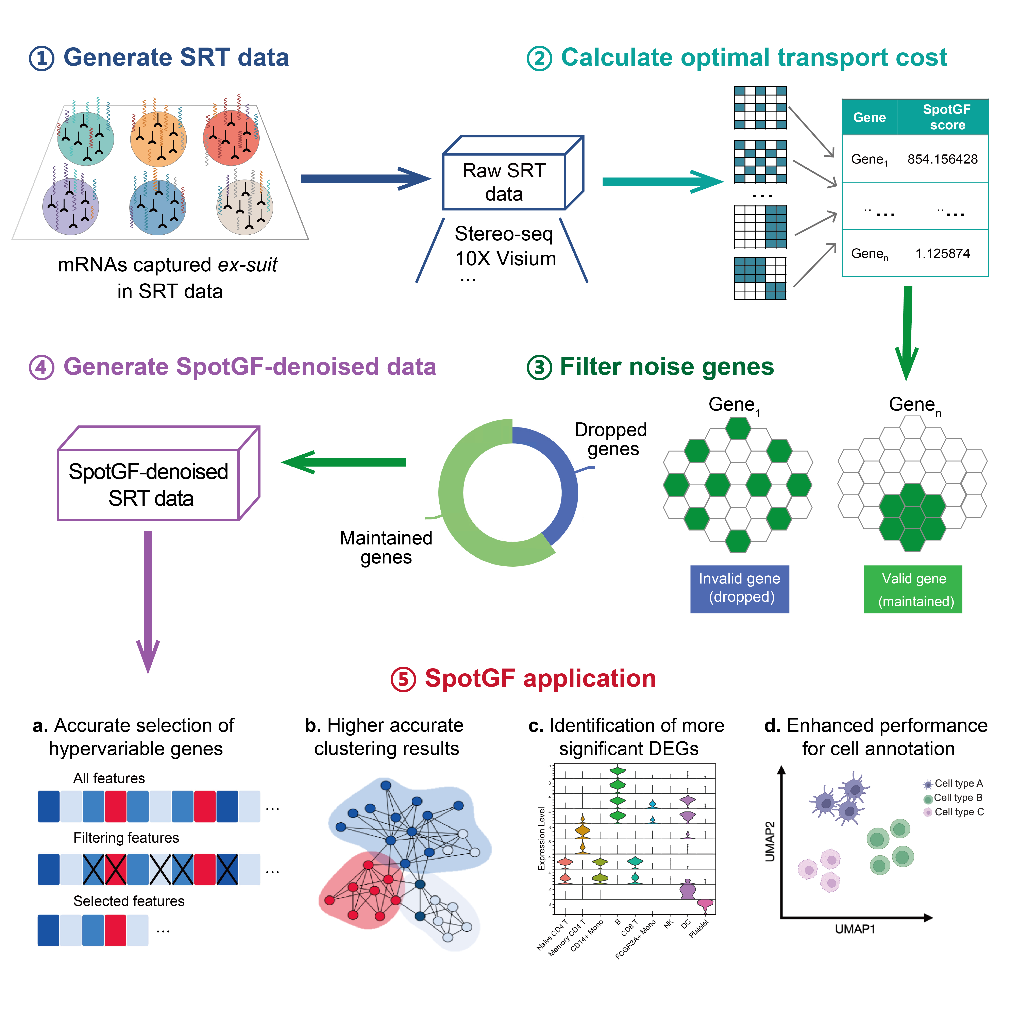 Principles and Applications of the SpotGF Denoising Algorithm
Principles and Applications of the SpotGF Denoising Algorithm
SpotGF has been tested across various datasets and shown to significantly reduce noise while enhancing the performance of data analysis tasks like cell clustering, cell type annotation, and identifying differentially expressed genes. These tasks are crucial for researchers trying to understand complex biological structures and the interactions within them.
For example, when applied to data from a study on soybean roots, SpotGF not only clarified which genes were actively expressed in specific cells but also improved the accuracy in identifying different cell types. This is particularly helpful for biologists looking to understand plant structures and their functions at a molecular level.
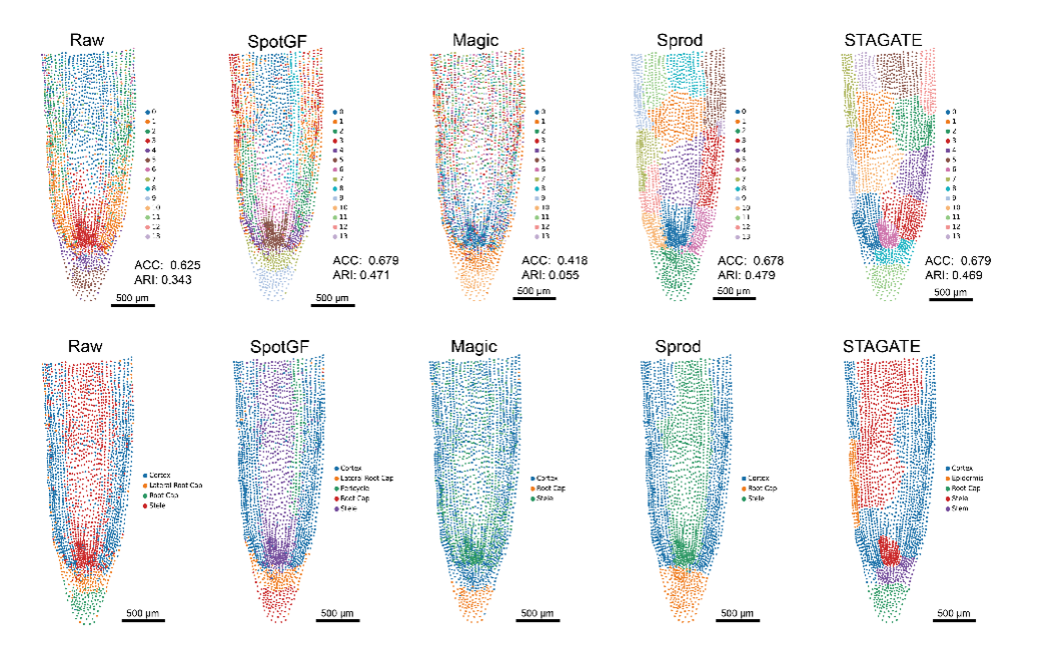 Performance Comparison of SpotGF and Existing Denoising Algorithms in Soybean Root Tip Stereo-seq Data
Performance Comparison of SpotGF and Existing Denoising Algorithms in Soybean Root Tip Stereo-seq Data
One of the major advantages of SpotGF is its user-friendliness and computational efficiency. Researchers can access SpotGF freely on GitHub, allowing them to apply this powerful tool to their own transcriptomic datasets. By streamlining the denoising process, SpotGF saves valuable time and resources, enabling scientists to focus more on their research questions rather than data cleaning.
The development of SpotGF is timely. As the use of spatially resolved transcriptomics grows, the demand for accurate data interpretation becomes even more critical. By effectively filtering out noise, SpotGF helps ensure that researchers can trust the spatial gene expression patterns they observe, leading to more reliable scientific discoveries.
This study can be accessed here: https://www.cell.com/cell-systems/abstract/S2405-4712(24)00269-2



