The development of multicellular organisms is an extraordinarily intricate process, orchestrated by the precise interplay of genes and cells across space and time. On June 26, researchers from BGI-Research, Southern University of Science and Technology and other institutes published a groundbreaking study in Cell. Using Drosophila melanogaster (fruit fly) as a model system, the researchers integrated advanced multi-omics sequencing technologies to decode the molecular mechanisms underlying development. The resulting single-cell 3D spatiotemporal multi-omics atlas, Flysta3D-v2, spans the entire development cycle of Drosophila from embryo to pupa. This comprehensive atlas provides unprecedented insights into cell type differentiation, spatiotemporal dynamics, and key regulatory networks, offering a transformative reference for developmental biology and a foundation for studying developmental disorders and related diseases.
 BGI Research and its partner institutes present Flysta3D-v2, offering unprecedented insights into cell type differentiation, spatiotemporal dynamics, and key regulatory networks of Drosophila melanogaster.
BGI Research and its partner institutes present Flysta3D-v2, offering unprecedented insights into cell type differentiation, spatiotemporal dynamics, and key regulatory networks of Drosophila melanogaster.
Flysta3D-v2 represents a breakthrough in 3D multi-omics mapping. The researchers utilized BGI’s proprietary Stereo-seq spatial transcriptomics technology, combined with single-cell RNA sequencing (scRNA-seq) and single-cell chromatin accessibility sequencing (scATAC-seq), to systematically sample key developmental stages of Drosophila. Samples were collected at intervals of 0.5-2 hours during embryogenesis, as well as from all larval and pupal stages. The team generated over 3.8 million spatially resolved single-cell transcriptomes and reconstructed high-precision 3D models using the Spateo algorithm. These models revealed intricate spatial dynamics of tissue morphology and gene expression with unprecedented accuracy, enabling the construction of a molecular timeline of Drosophila development.
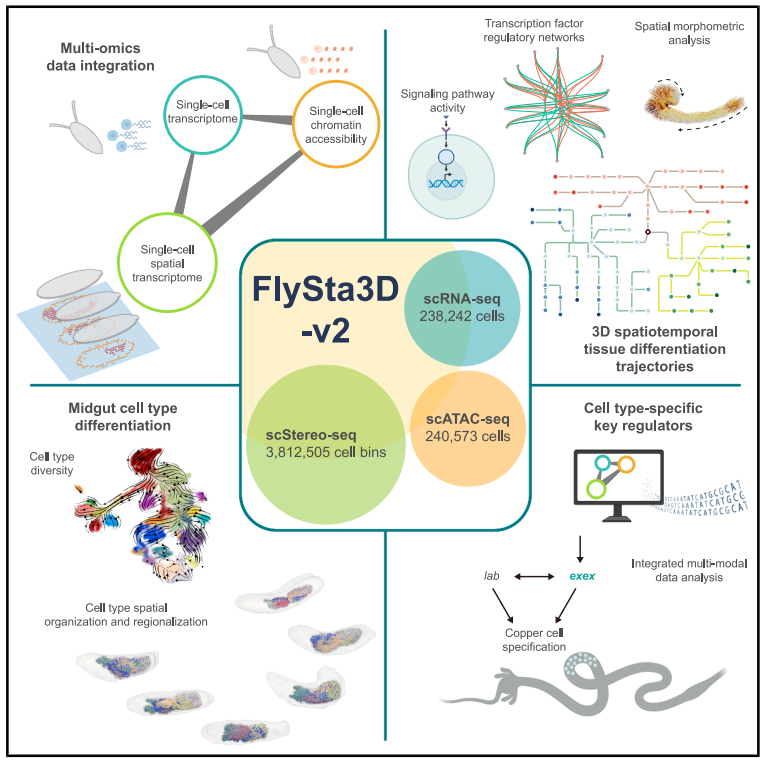 Graphical abstract of the study.
Graphical abstract of the study.
A Molecular Clock for Decoding Development
Integrating spatial transcriptomics, single-cell transcriptomics, and chromatin accessibility data, the researchers identified continuous differentiation trajectories of embryonic cells, offering a spatiotemporal blueprint of cell fate decisions. Using advanced algorithms, they aligned disparate datasets to a unified developmental timeline, uncovering key transcription factors (TFs) that govern cell fate determination within each germ layer. Furthermore, they discovered previously uncharacterized TFs with potential tissue-specific functions in the nervous, cardiac, and endocrine systems, expanding the understanding of developmental regulation.
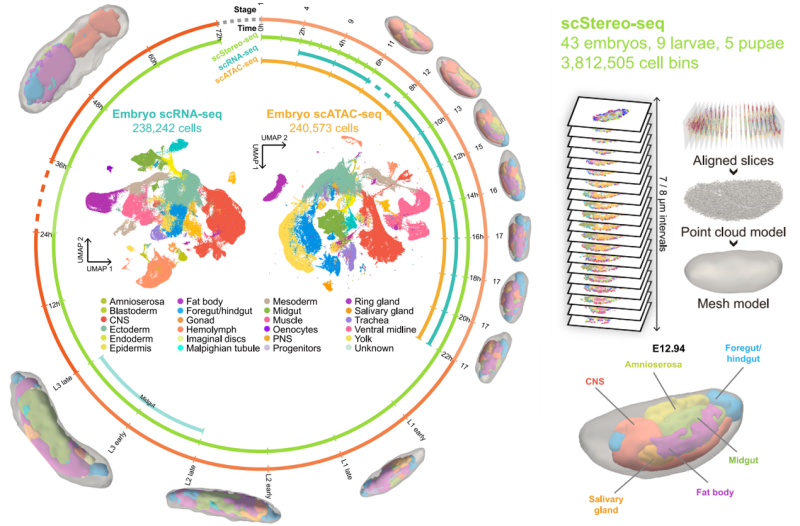 The single-cell spatiotemporal multi-omics atlas of developing Drosophila, which maps the entire developmental process while serving as a "molecular clock" to decode how cells differentiate, interact, and organize over time and space.
The single-cell spatiotemporal multi-omics atlas of developing Drosophila, which maps the entire developmental process while serving as a "molecular clock" to decode how cells differentiate, interact, and organize over time and space.
Revealing Patterns of Tissue Differentiation and Neural Dynamics
The atlas also revealed critical insights into tissue differentiation and neural dynamics. For the first time, the spatial origins of tissue differentiation were mapped at the single-cell level. Using the fat body, a Drosophila organ homologous to the mammalian liver, and the foregut and hindgut, which correspond to the stomach and large intestine, the researchers uncovered distinct differentiation patterns. The fat body exhibited a decentralized mode of differentiation, with spatially intermixed cell types reflecting the scattered distribution of precursor cells during early embryogenesis. In contrast, the foregut and hindgut displayed centralized differentiation, with spatially clustered cell types at different developmental stages. This suggests the presence of specific stem cell niches and provides new evidence for understanding gut morphogenesis.
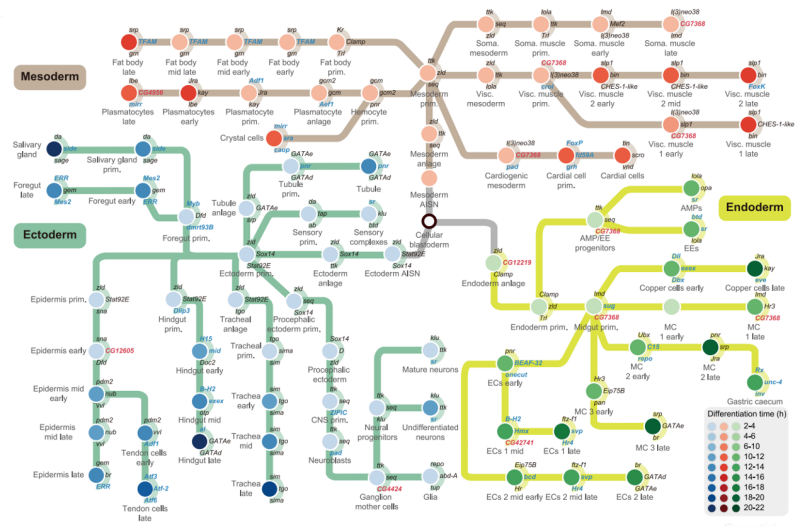 Metro plot showing tissue differentiation trajectories based on cluster phylogeny for major tissues of three germ layers.
Metro plot showing tissue differentiation trajectories based on cluster phylogeny for major tissues of three germ layers.
The study also tracked the development of the central nervous system (CNS) using Flysta3D-v2. Morphometric analyses revealed that the posterior ventral nerve cord (VNC) and anterior brain regions drive distinct phases of CNS remodeling. Early-stage changes were found to depend on cell fate cell fate decisions, while later stages were associated with the activation of signaling pathways. The research also identified novel regulators of CNS development, such as CG42394, a transmembrane protein, and lncRNA:CR30009, a long non-coding RNA. Both were shown to be associated with neuroblast migration, and experiments confirmed their CNS-specific expression, highlighting their potential roles in neural development.
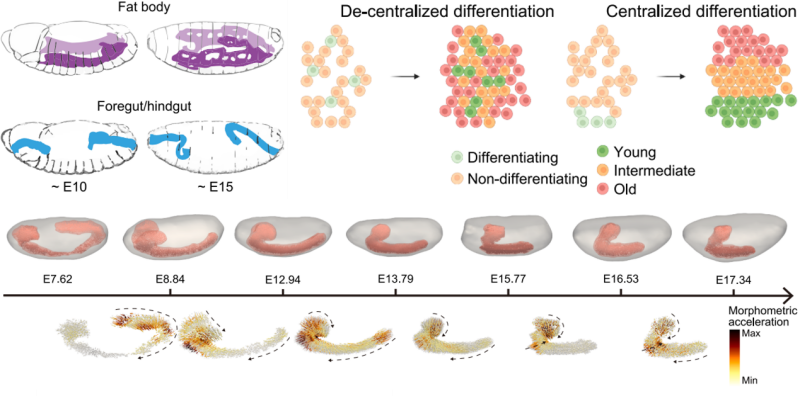 A schematic representation of the anatomical morphology of the developing fat body and foregut/hindgut (left) and the centralized versus decentralized differentiation modes (right).
A schematic representation of the anatomical morphology of the developing fat body and foregut/hindgut (left) and the centralized versus decentralized differentiation modes (right).
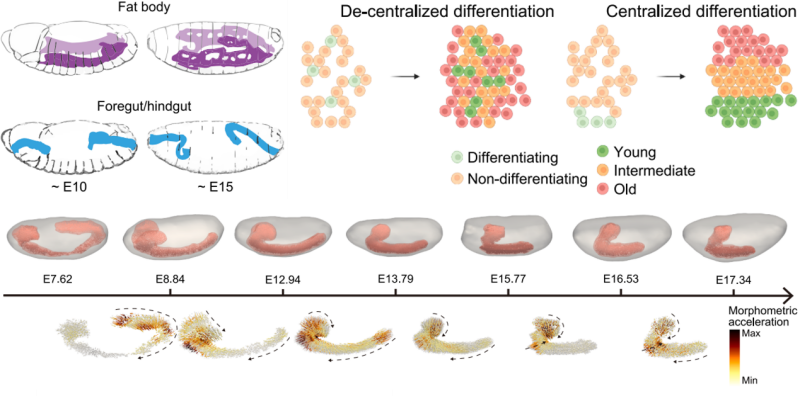 3D models of CNS, CNS cell migration trajectories, and acceleration scores across 7 scStereo-seq samples of developmental age between 7 and 18 h.
3D models of CNS, CNS cell migration trajectories, and acceleration scores across 7 scStereo-seq samples of developmental age between 7 and 18 h.
Tracing the Spatiotemporal Evolution of the Midgut
In addition to uncovering neural dynamics, the study provided a detailed view of midgut development. Using scStereo-seq (single-cell Stereo-seq), the researchers mapped the dynamic changes in midgut cell types and spatial organization during larval and pupal stages.
The larval midgut was found to undergo significant growth and diversification, with the differentiation of entero-endocrine cells (EEs, responsible for stimuli sensing and hormone secretion) into distinct subclusters that exhibited unique spatial distributions along the anterior-posterior axis. These patterns mirrored the functional compartmentalization of the adult midgut.
During the late larval stage (L3), dramatic reductions in specific cell types, such as anterior gastric caecum cells, were observed. This coincided with upregulation of autophagy-related genes, confirming the role of programmed cell death in midgut remodeling before metamorphosis.
In the pupal stage, the midgut differentiated into inner and outer layers, each with distinct gene expression profiles. The inner layer was enriched in genes related to metal ion metabolism, while the outer layer expressed antimicrobial genes, suggesting specialized roles during metamorphosis.
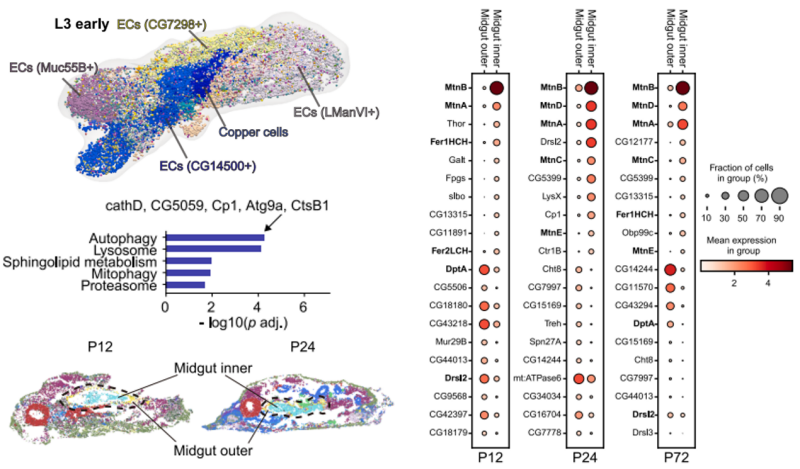 Cell-type diversity and functional regionalization in developing midgut.
Cell-type diversity and functional regionalization in developing midgut.
The study further revealed that midgut functional regionalization begins as early as the embryonic stage. Gene expression patterns corresponding to adult midgut regions (R1-R5) gradually emerged along the anterior-posterior axis during late embryogenesis. Regional differences were also observed in adult midgut progenitors (AMPs) during larval development, providing a developmental basis for region-specific regeneration in adulthood.
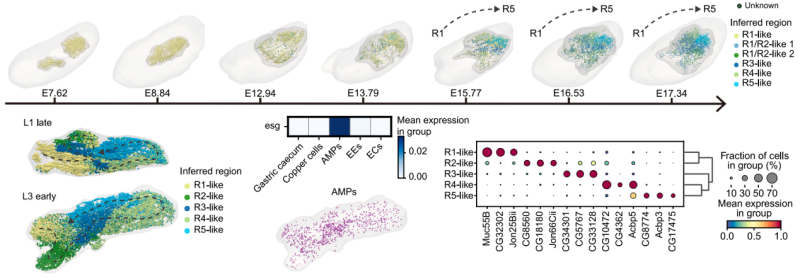 3D embryo and midgut models across representative embryo scStereo-seq samples, showing spatial distribution of inferred midgut regions.
3D embryo and midgut models across representative embryo scStereo-seq samples, showing spatial distribution of inferred midgut regions.
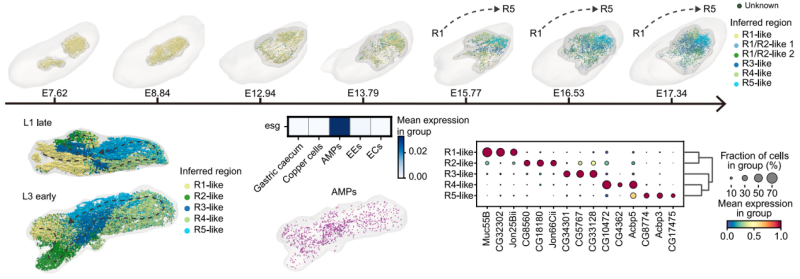 Same as above but for larva scStereo-seq samples, showing only midgut models, and dashed lines indicate inferred midgut backbones.
Same as above but for larva scStereo-seq samples, showing only midgut models, and dashed lines indicate inferred midgut backbones.
Key Regulator exex Identified in Copper Cell Development
One of the most significant discoveries of the study was the identification of exex, a transcription factor critical for the development of copper cells in the midgut.
Copper cells, which are essential for maintaining metal ion homeostasis and gastric acid secretion, were shown to be regulated by exex in a spatiotemporally specific manner. The researchers found that exex is highly expressed during the early differentiation of midgut progenitors into copper cells. Although its expression decreases in mature copper cells, exex binding motifs remain active, continuously regulating key target genes.
Knockdown experiments demonstrated that exex is essential for copper cell differentiation and function. Loss of exex led to a sharp reduction in copper cell numbers, abnormal cell morphology, and impaired copper ion accumulation.
The study also revealed that exex regulates copper cell development through a network involving the transcription factors kay and lab, highlighting a co-regulatory mechanism that enhances gene activation.
In May 2022, an international team of scientists led by BGI-Research utilized Stereo-seq technology to create the world’s first panoramic spatial atlases of life. These groundbreaking findings were published in Cell as part of the SpatioTemporal Omics Consortium (STOC) article collections, with the Drosophila developmental atlas highlighted in Developmental Cell.
Dr. Longqi Liu, a researcher at BGI-Research and co-corresponding author, stated: "This study builds upon our previous work on the Drosophila atlas, achieving single-cell resolution and enriching it with additional omics data, such as single-cell transcriptomes. As a result, we have constructed the first 3D multi-omics atlas spanning the entire developmental cycle of Drosophila – a classic model organism – providing molecular-level insights into the mechanisms of plant development. Moreover, by integrating multiple omics technologies, our research team has systematically described the developmental process of Drosophila using a simpler and more efficient approach, offering a reference framework for studying the development of other organisms, including humans."
The dataset is publicly available through the Flysta3D-v2 database (https://db.cngb.org/stomics/flysta3d-v2/), which offers interactive 3D visualizations of gene expression patterns, regulatory networks, and signaling pathways. This resource is poised to drive advances in developmental biology, providing new insights into the molecular basis of life’s spatiotemporal complexity.
Ethical approval for this study was obtained. The full study can be accessed here: https://www.cell.com/cell/abstract/S0092-8674(25)00629-4



