Researchers from the 10,000 Fish Genomes Project (Fish10K) consortium report a comprehensive genomic resource spanning 464 teleost fish species, including 110 newly assembled genomes that fill three previously unsampled orders. The study, accepted by The Innovation (IF=25.7), brings fish comparative genomics to the scale previously achieved in birds and mammals and provides a genome-wide view of teleost evolution over ~300 million years. By generating a reference-free, 464-way whole-genome alignment, the authors offer a consolidated framework to revisit long-standing questions in fish evolution, from the timing of major radiations to the genetic elements that shape fins, brains, hearts and gills.
The study “A genomic compendium of hundreds of teleost fishes reveals their evolutionary landscape” was published in The Innovation.
Logo of the 10,000 Fish Genomes Project (Fish10K).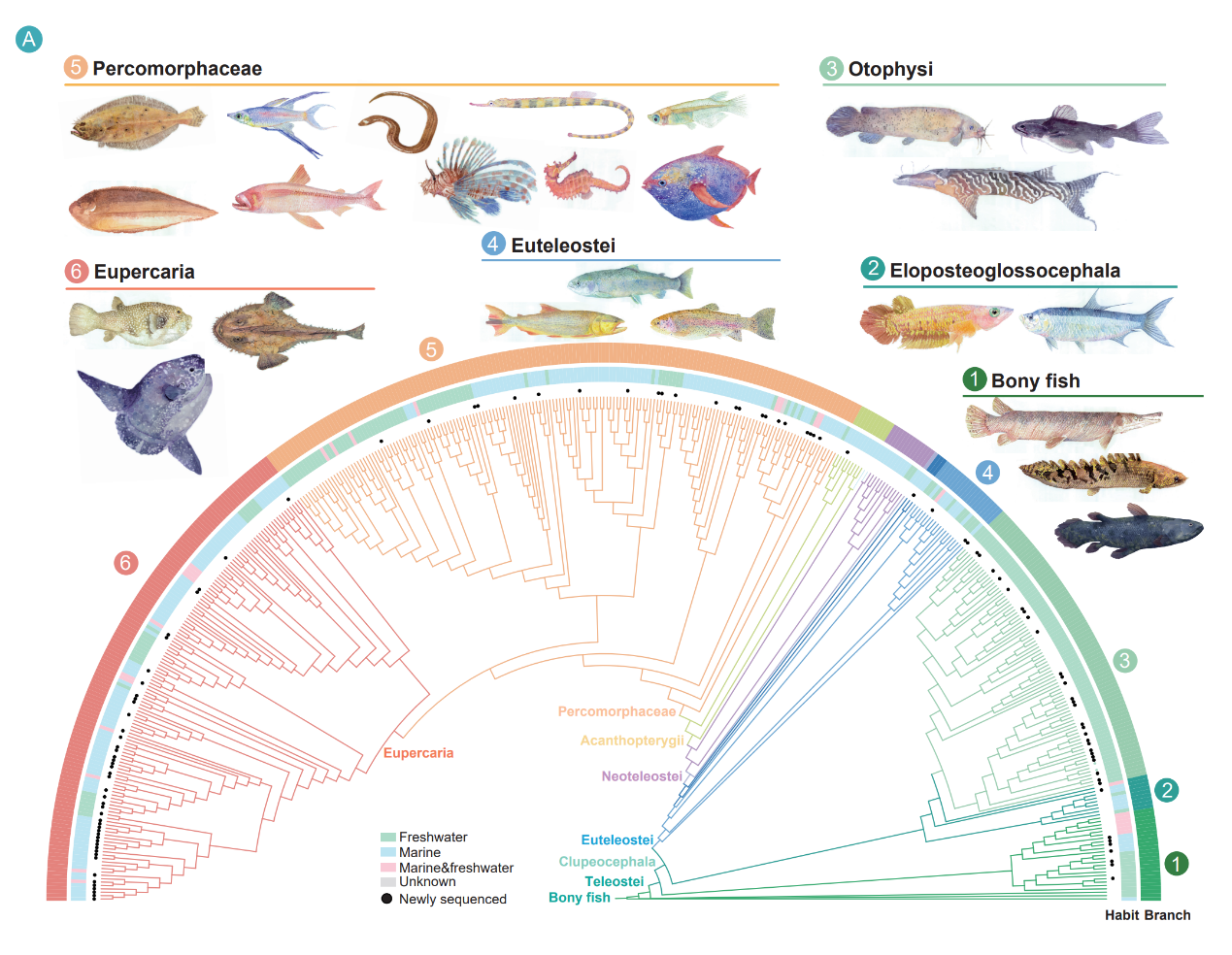
The 464-species phylogeny with 110 newly sequenced genomes establishes the largest fish-wide comparative genomics resource covering all teleost orders, enabling systematic studies across teleost diversity for ecology, aquaculture and conservation research.
The whole-genome analysis challenges the widely accepted "Siluriphysi" grouping, instead supporting Gymnotiformes as sister to a Characiformes–Siluriformes clade rather than forming a unique pair with catfishes. This topology reframes how electroreception may have evolved within these lineages and highlights the power of genome-wide data to resolve contentious phylogenetic relationships.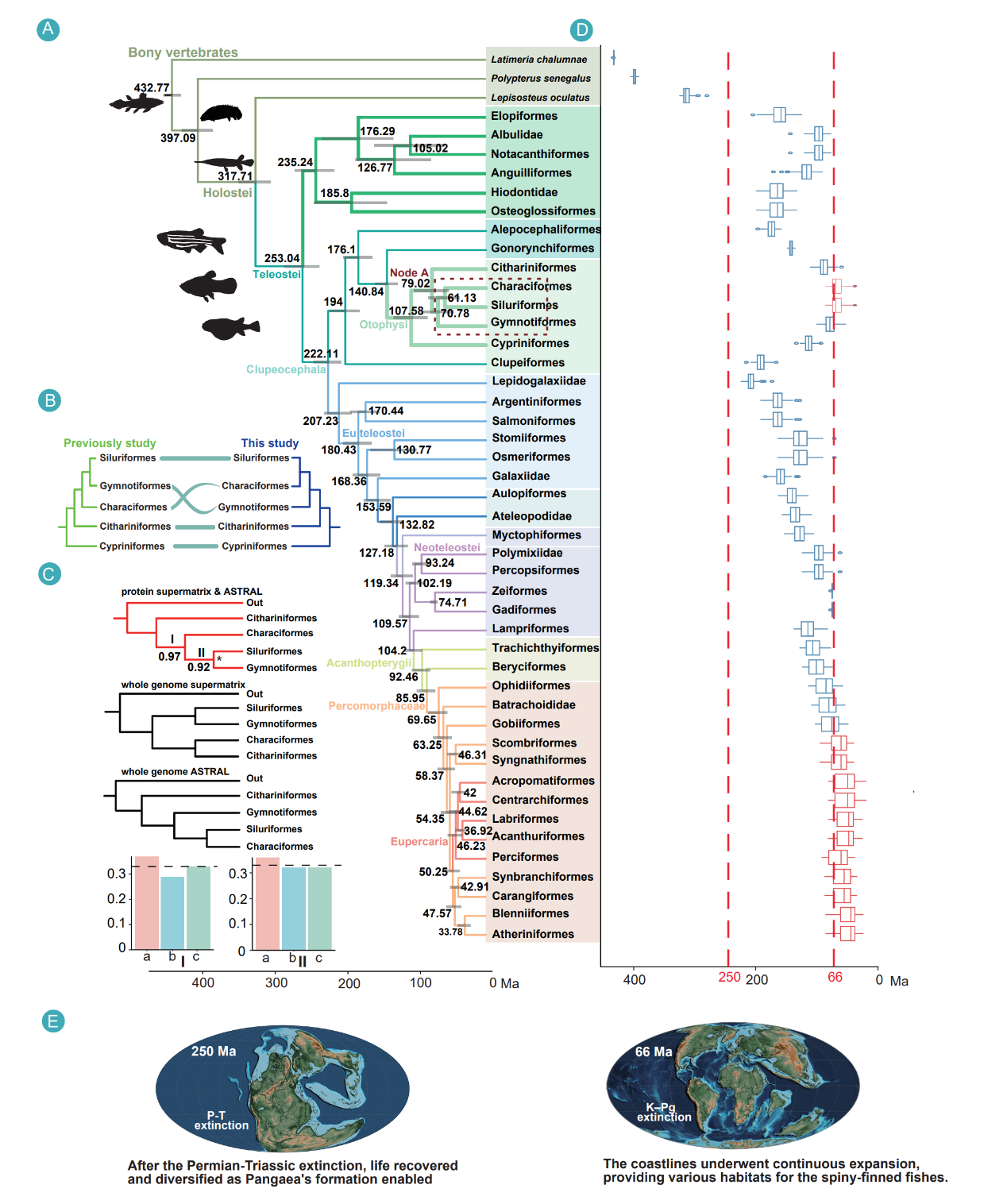
Whole-genome phylogenomics challenges the widely accepted "Siluriphysi" monophyly and dates crown teleost origins to ~253 Ma, resolving long-standing evolutionary debates and reframing how electroreception evolved, which helps researchers understand the genetic basis of sensory adaptations in fishes.
The timeline places crown teleost origins at approximately 253 million years ago, slightly predating the end-Permian mass extinction. Rather than a simple, continuous acceleration after the Cretaceous–Paleogene boundary, the analysis reveals three phases of diversification: an initial rapid burst, a longer period of declining rates through the Jurassic–Cretaceous, and a post-K–Pg phase that stabilizes with only transient rebounds. The study further observes that episodes of global warming coincide with suppressed diversification, adding environmental context to macroevolutionary dynamics.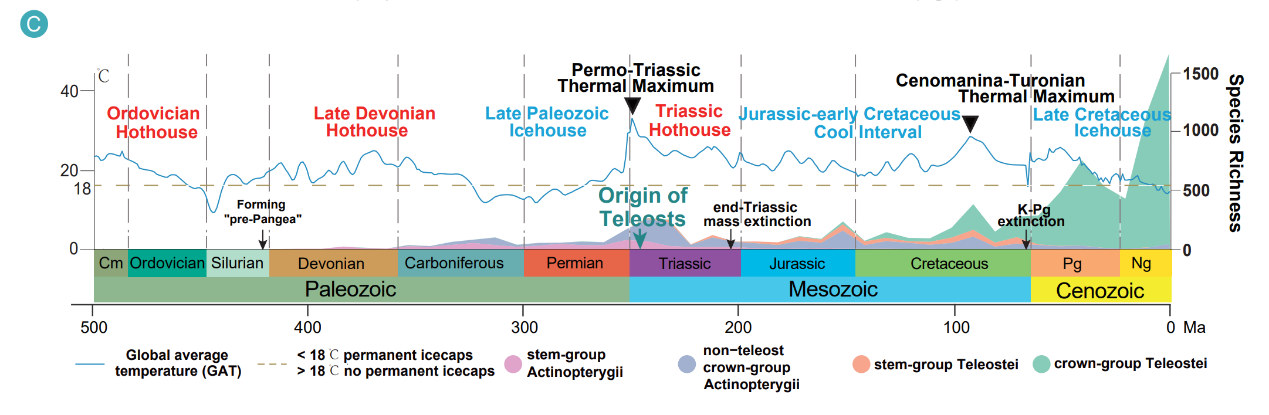
Fossil evidence reveals that global warming episodes suppressed teleost diversification, providing critical insights into how climate change impacts species diversity that can inform conservation strategies in the face of current warming.
At the level of genome architecture, the compendium highlights a distinct transposable-element landscape. Teleost genomes are characterized by DNA transposons as the predominant class, in contrast to the LINE-dominant profiles in other vertebrates. Freshwater teleosts carry significantly higher transposon content than marine species, suggesting that mobile DNA has contributed structural and regulatory raw material as teleosts adapted to shifting osmotic and ecological regimes.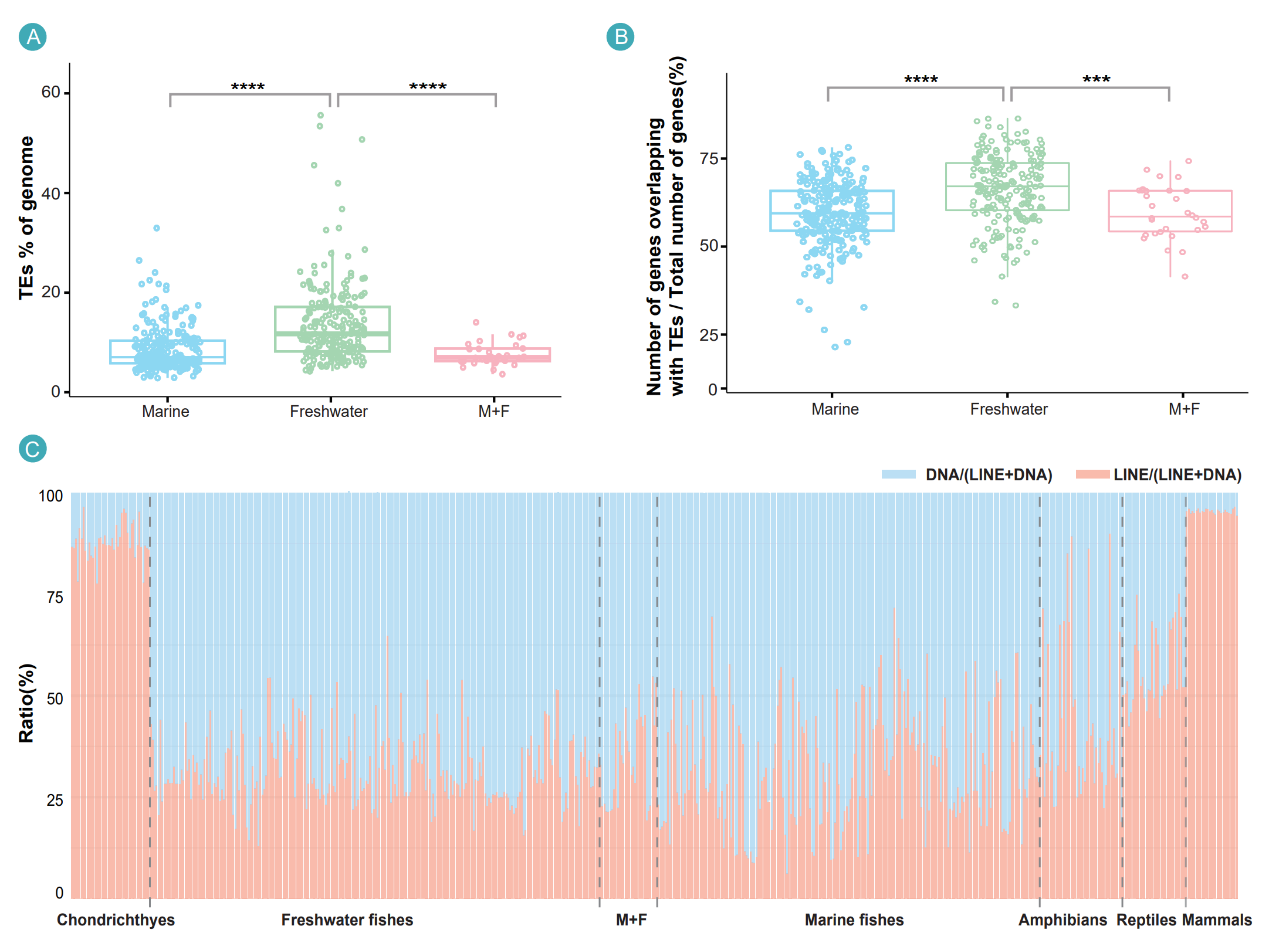
Freshwater teleosts harbor higher transposon content than marine species, and teleost genomes uniquely feature DNA-transposon dominance rather than the LINE-dominant pattern seen in other vertebrates, revealing how mobile DNA may have contributed to environmental adaptation and teleos radiation.
The whole-genome alignment enables a comprehensive survey of constraint and acceleration across the fish genome, identifying constrained sequences enriched in non-coding regions and linked to developmental processes. The authors catalog teleost highly conserved elements and teleost-specific conserved elements that are widely conserved across teleosts but absent from holosteans. Notably, teleost-specific conserved non-coding elements are enriched near genes retained from the teleost-specific whole-genome duplication, suggesting that duplication not only expanded coding repertoires but also created genomic contexts conducive to new regulatory elements. These elements overlap open chromatin and bear regulatory marks, indicating potential roles as cis-regulatory sequences that may fine-tune expression relevant to teleost-specific traits.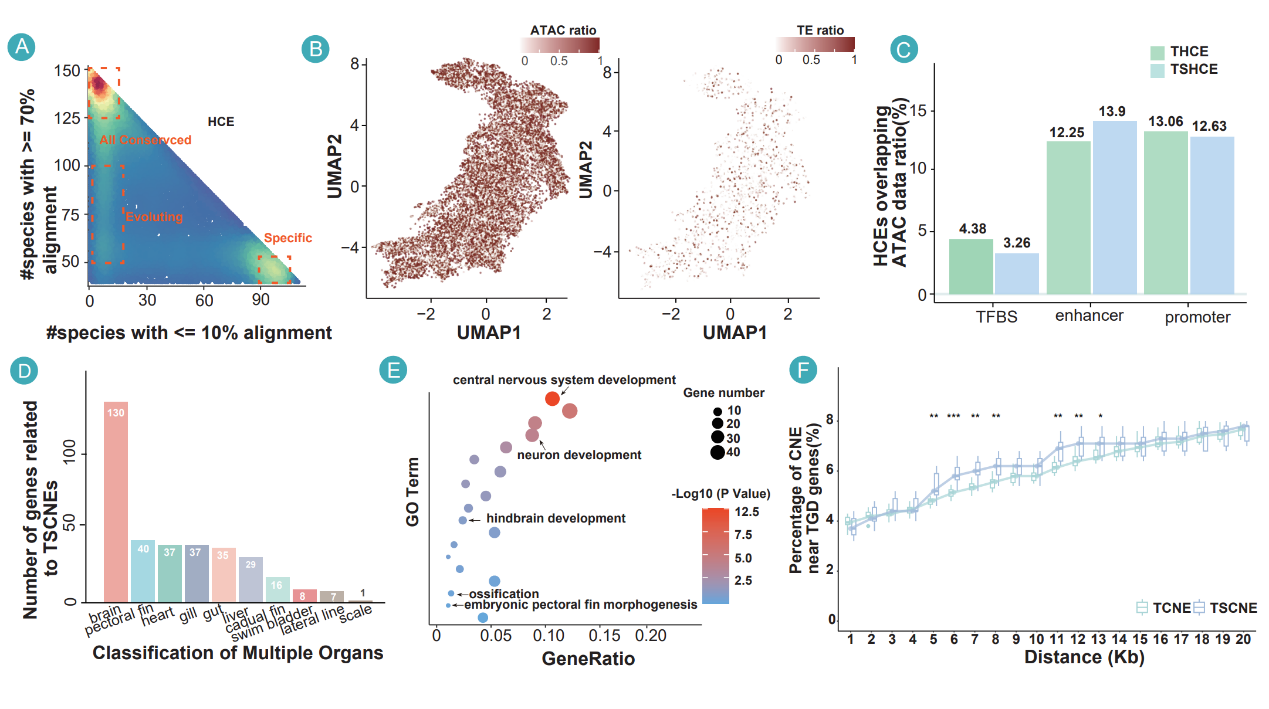
Teleost-specific conserved elements enriched near whole-genome duplication-derived genes demonstrate that duplication facilitated both coding and regulatory innovation, providing candidate regulatory regions for understanding teleost-specific traits like fin morphogenesis and neural development.
Together, these results establish a fish-wide, alignment-centric resource that can support comparative and functional studies across ecology, aquaculture and model-organism biology. By supplying both a solid backbone phylogeny and a map of conserved and accelerated elements, the compendium can help researchers trace trait evolution across teleost diversity, prioritize candidate regulatory regions in zebrafish and other models, and provide genomic context for species of conservation and economic importance. The authors have deposited the data in public repositories, with scripts available via the Fish10K GitHub repository to facilitate reuse by the community.
All experimental animal treatments were verified and identified by taxonomic experts and museum taxonomists under protocols.
This research can be accessed here: https://doi.org/10.1016/j.xinn.2025.101177



