In the latest issue of Nature Genetics, BGI-Research, in collaboration with Northwestern Polytechnical University, China, the University of Otago, New Zealand, the New Zealand Institute for Plant and Food Research, and other institutes, reported the most detailed and highest-quality genome assemblies to date for three Allium species: onion, garlic, and Welsh onion, as well as their close relative, the African lily. Using BGI Group's spatial multi-omics technology Stereo-seq, the study unveiled the development, metabolism, cell-type categorization, and gene expression changes during different stages of development in the Allium species. As a project under the Earth BioGenome Project (EBP), these results provide profound insights into understanding these ancient and valuable species and improving their cultivation and breeding.
 The research ‘Chromosome-level genomes of three key Allium crops and their trait evolution’ published in Nature Genetics.
The research ‘Chromosome-level genomes of three key Allium crops and their trait evolution’ published in Nature Genetics.
Allium is a genus of monocotyledonous flowering plants with hundreds of species, including cultivated onion, garlic, scallion, shallot, leek, Welsh onion, and chives. The earliest records of humans using onions as food date back to Egyptian tombs from 3,000 BC. Today, onions are second only to tomatoes as one of the most cultivated vegetables globally, with 100 million tons of dried onions produced annually.
Despite their vital role on the dining table, questions about Allium species, such as their evolution and the source of their flavor, have puzzled the world for centuries due to the lack of high-quality reference genomes.
In this research, scientists have assembled the most complete genomes for onion, garlic, Welsh onion, and African lily to date, with the highest quality. The contig N50 sizes of the three Allium species (onion, garlic, and Welsh onion) were not only the highest among recently published Allium assemblies but also the highest recorded for plants with large genomes.
N50 statistics are often used to describe the assembly quality or the 'completeness' of a genome assembly in terms of contiguity. A contig is a set of DNA segments that overlap in a way that provides a contiguous representation of a genomic region. Given a set of contigs, the N50 is defined as the sequence length of the shortest contig at 50% of the total assembly length. A larger contig N50 size indicates that the genome assembly is closer to the original genome.
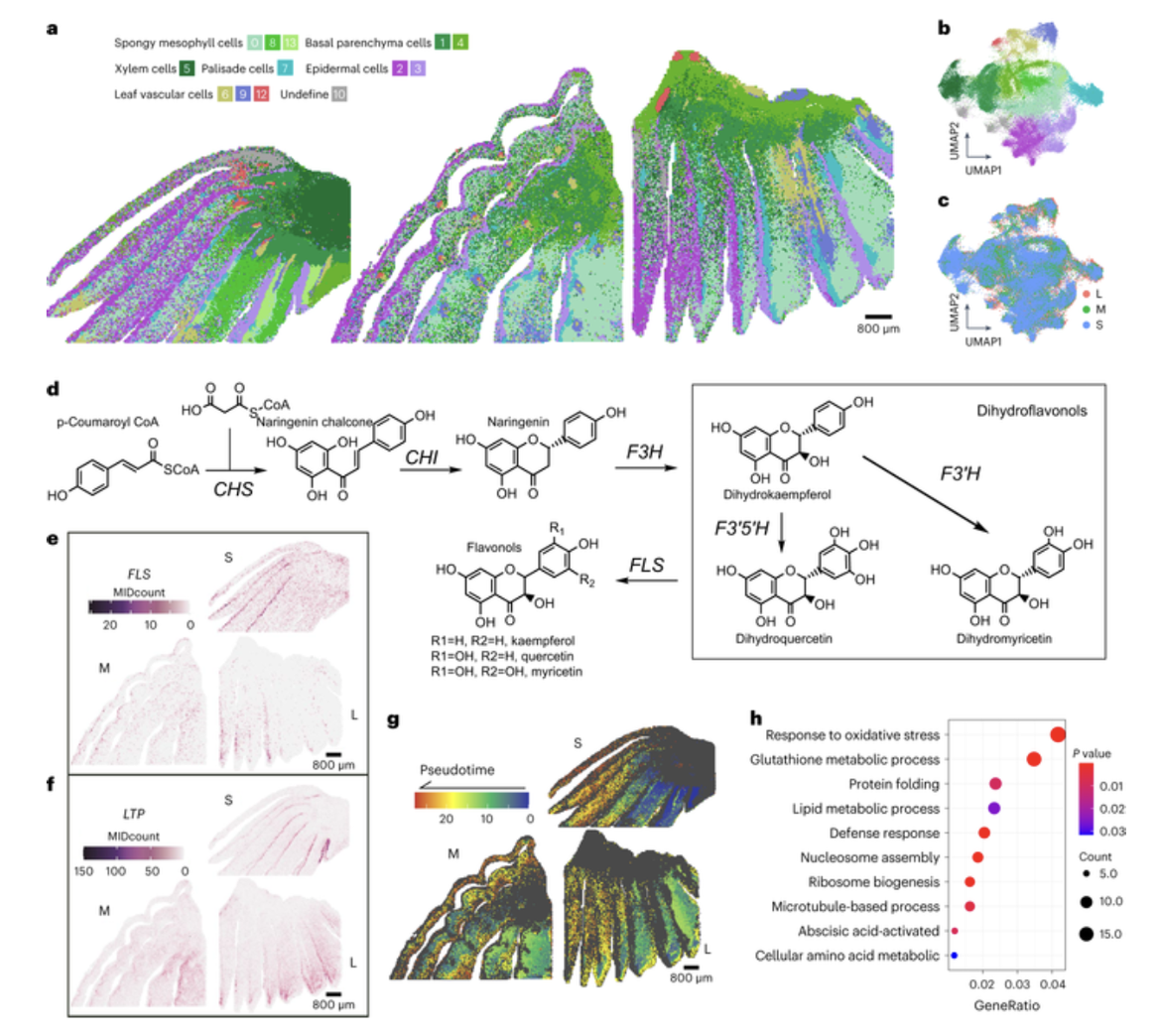
Stereo-seq technology played a pivotal role in this study by enabling the analysis of the spatiotemporal transcriptomes of onion bulbs. This analysis unveiled variations and commonalities in the expression patterns of genes associated with flavonoid synthesis across diverse cell types, the attributes of gene expression involved in epidermal wax synthesis, and the dynamics of spongy mesophyll cell development, among other insights.
The research team selected onions at early, intermediate, and mature growth stages and conducted Stereo-seq sequencing on samples taken from the bulb bases. Onions are biennial plants featuring large bulb storage organs for winter survival. These bulbs harbor a variety of secondary metabolites, including flavonoids like quercetin, a prominent flavonoid in onions known for its antioxidant properties and disease resistance.
 Stereo-seq technology reveals spatial distribution of gene expression in metabolic process and onion bulb formation.
Stereo-seq technology reveals spatial distribution of gene expression in metabolic process and onion bulb formation.
Previous studies have identified a group of genes responsible for flavonoid synthesis in onion bulbs. The Stereo-seq results in this study revealed that these genes, with the exception of one, are co-expressed in epidermal cells. This implies that epidermal cells in onion bulbs serve as the primary site for flavonoid synthesis, indicating that they are the primary source of flavonoids in onion bulbs.
Furthermore, the onion epidermis exhibited high expression levels of a specific gene family involved in the formation of onion epidermal wax, a skin-like layer that safeguards the underlying delicate tissues from harmful viruses and fungi. These findings provide crucial insights for future research on onion pest control and the development of pest-resistant onion varieties.
Researchers also delved into the process of onion bulb formation, a well-organized spatiotemporal change in cell development. Based on spatial transcriptome analysis and previous studies, bulb formation appears to entail several cellular processes. During the development of onion bulbs, spongy mesophyll cells, a loosely arranged cell layer in the inner regions of onion bulb layers responsible for gas exchange and the regulation of gas flow, followed a continuous trajectory of cell formation from the inner to the outer layer and from the base to the top. In each layer of the bulb, the expansion of sponge mesophyll cells occurred from the outer edge to the inner edge, indicating that the innermost cells began expansion earlier and were subsequently pushed further inward by the later expansion of cells near the outer edge.
"Based on high-resolution Stereo-seq technology, our data provide new insights into onion bulb development and metabolism even at the subcellular nanoscale, with 100-fold higher resolution than previous spatial technology," said Dr. Xu Xun, Director of BGI-Research. "In the increasingly severe context of global biodiversity loss, this study highlights the significant value of utilizing high-precision genomic data for genetic analysis and research on biodiversity resources."
Read the research article: https://www.nature.com/articles/s41588.023-01546-0



