Shenzhen, September 4 - In a landmark study published in Nature, an international team of researchers led by BGI-Research has reported the most comprehensive global marine microbiome database to date, which unveiled the immense potential of marine microorganisms for marine science research, and biotechnological and biomedical applications.
 The study “Global marine microbial diversity and its potential in bioprospecting” was published in Nature.
The study “Global marine microbial diversity and its potential in bioprospecting” was published in Nature.
Marine microorganisms are an important part of the marine ecosystem and produce many active substances and enzyme resources with novel structures and unique functions. They hold significant potential for applications in the fields of new marine medicines, biomaterials, biocatalysis, and functional foods. Over the past two decades, the number of microbial genomes retrieved from marine environments has significantly increased. However, translating this marine genomic diversity into biotechnological and biomedical applications has been challenging.
The study was led by BGI-Research, in collaboration with an international team of scientists, including researchers from Shandong University, the University of East Anglia (UK), Ocean University of China, Xiamen University, the University of Copenhagen (Denmark), and other institutions.
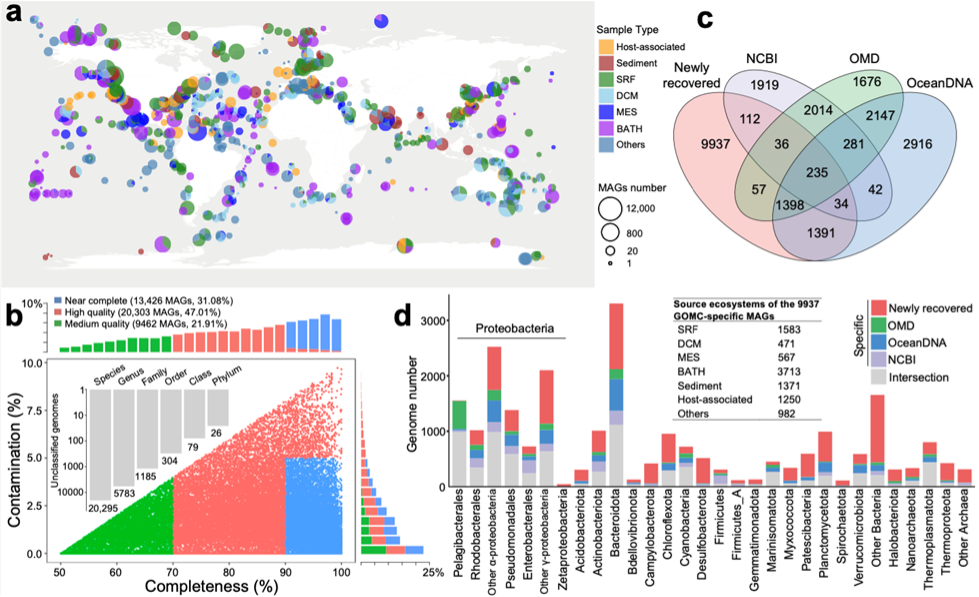
Using metagenomic binning approach, the research team spent five years reanalyzing nearly 240 terabytes of publicly available marine metagenomic data from the National Center for Biotechnology Information (NCBI), European Bioinformatics Institute (EBI), and Joint Genome Institute (JGI), covering the period from August 2009 to July 2020. This effort generated a database containing 43,191 metagenome-assembled genomes (MAGs) and 2.46 billion gene sequences, spanning 3,470 microbial genera and across 138 phyla. Notably, more than 20,000 of these genomes (46.99%) were identified as potential novel species.
Combining these MAGs with public marine bacterial and archaeal genomes from NCBI, Ocean Microbiomics Database (OMD) and OceanDNA, the research team constructed a unified global ocean microbiome genome catalogue (GOMC), which represents the most comprehensive global marine microbiome database to date. The GOMC reports nearly 10,000 new discovered genomes from deep-sea, sediment, symbiotic and other habitats.
 An overview of the global ocean microbiome database.
An overview of the global ocean microbiome database.
The study has remarkably expanded the known diversity of taxa such as the marine archaea Thermoproteota and Halobacteriota, as well as the marine bacteria Campylobacteta and Desulfobacterota. It refreshes the upper limit of the genome size of marine prokaryotic microorganisms, and also explains potential mechanisms for the observed microbial genome expansion, such as the selective pressure of fluctuating anoxic marine environment on the bacterial evolution of large and complex genomes.
At the same time, the biogeographic distribution pattern of marine microbial communities around the global ocean was calculated, providing a new perspective for understanding how microorganisms are distributed and maintained in different environments. The research will provide unprecedented opportunities for the evolution, environmental adaptation and ecology of marine microorganisms.
The research team explored the genetic resources of the GOMC database from multiple dimensions, leading to the identification of 36 CRISPR-Cas9 systems. Researchers further investigated the application potentials of one of these systems, Om1Cas9. The results demonstrated that Om1Cas9 is highly effective for genome editing in all testing scenarios, highlighting its potential as a new tool for cancer research and treatment development.
A total of 117 potential new antimicrobial peptides were identified using deep learning-based bioinformatics method, among which the antimicrobial peptide cAMP_87 exhibited excellent antimicrobial activity against both Gram-negative and Gram-positive bacteria. This discovery could pave the way for the future development of alternatives to existing broad-spectrum antibiotics, as well as skincare products, livestock feeding, and other areas.
 The comparison of the plastic-degrading capabilities of land- and marine-based PET-degrading enzymes.
The comparison of the plastic-degrading capabilities of land- and marine-based PET-degrading enzymes.
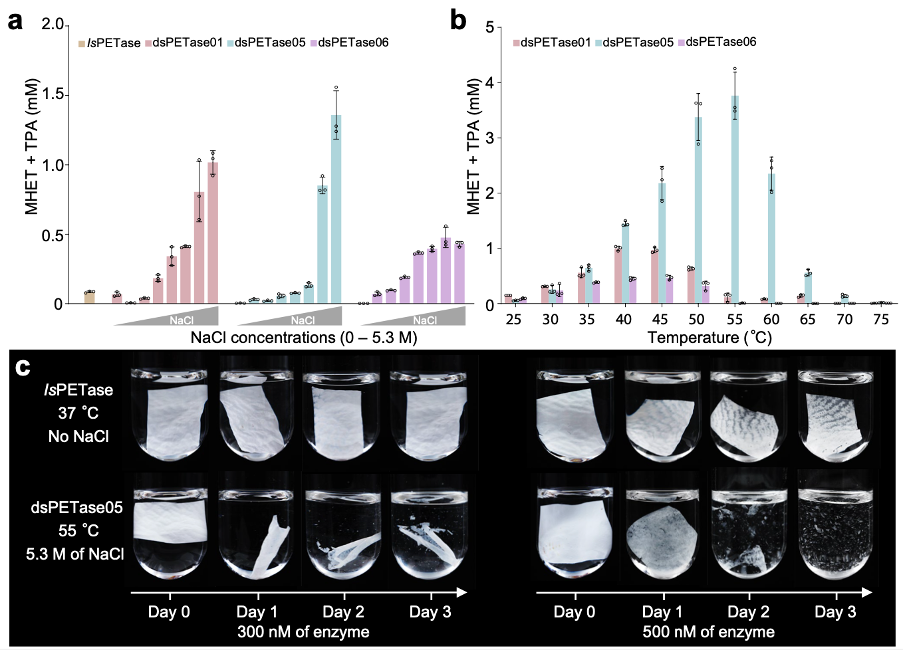
Several new polyethylene terephthalate (PET) plastic-degrading enzymes (dsPETase05, 06, 01) with high activity potential were also discovered. Among which, dsPETase05 can degrade PET plastic film within three days, achieving a degradation rate of 83%, which is more than four times the efficiency of the PET-degrading enzyme (IsPETase) discovered in land microorganisms. These research findings are expected to drive application developments in related industries and potentially revolutionize current PET plastic handling methods.
Dr. Chen Jianwei, co-first author of the study and a researcher from BGI-Research, commented, “This research marks a major step forward in marine metagenomics, showing how important marine microorganism are for improving human health and supporting environmental sustainability. These findings open up promising opportunities for scientists worldwide to explore and use ocean resources sustainably, while also setting the stage for future breakthroughs in biotechnology and biomedicine.”
Marine Microbiology Professor Thomas Mock from the University of East Anglia’s School of Environmental Sciences and co-corresponding author of the study said that the work takes the field of ocean metagenomics to the “next level.” “While previous studies have provided initial insights into the role of marine systems in maintaining biological diversity, our research not only builds on these findings but also introduces new opportunities for sustainable exploration and utilization of the oceans, which is overdue considering the global challenges the human society faces on our planet,” he said.
This study can be accessed here: https://www.nature.com/articles/s41586-024-07891-2
The Global Ocean Microbiome Catalogue can be accessed here: https://db.cngb.org/maya/datasets/MDB0000002



