Intracerebral hemorrhage (ICH) may account for barely one in seven strokes, yet it is the deadliest form: more than forty percent of patients die within days and many survivors never walk or speak independently again. Clinicians know that blood leaking into the brain sets off waves of inflammation, scarring and, occasionally, repair. however, a high-resolution, time-coded map of the processes occurring within injured tissue has been lacking.
Published in Neuron, scientists from BGI-Research, the School of Basic Medical Sciences at Zhengzhou University, the Karolinska Institute and the University of Gothenburg now provide exactly that guide. Using BGI's proprietary spatial multi-omics technology, Stereo-seq - a breakthrough spatial transcriptomics platform that reads every RNA molecule in situ at subcellular resolution - the team followed more than 3.3 million individual cells in mouse brains from three hours to twenty-eight days after an experimental bleed, piecing together the first spatiotemporal transcriptomic maps of ICH at single-cell resolution.
 The study “Spatiotemporal transcriptomic maps of mouse intracerebral hemorrhage at single-cell resolution” was published in Neuron.
The study “Spatiotemporal transcriptomic maps of mouse intracerebral hemorrhage at single-cell resolution” was published in Neuron.
The freely accessible dataset via the STOmics portal reveals the precise timing of neuronal death, the movement of immune and glial cells to clear debris, the hardening of scar tissue, and the reactivation of genes for myelin and axon regeneration at the lesion rim days later.
The atlas demonstrates that the clot created a molecularly distinct core within hours; damaged neurons linger briefly but are replaced almost entirely by macrophages, microglia and a dense sleeve of reactive astrocytes by day seven, effectively isolating the injury from healthier tissue. Four concentric molecular zones emerge at that time point: the innermost expresses heme and iron-handling genes such as Hmox1 and ferritin, the next upregulates matrix-remodeling enzymes and growth factors, the third layer bristles with the scar protein GFAP, and the outermost shows revived expression of Nfasc, Cnp and Sirt2, indicating axonal regrowth and remyelination. That clean-up, rebuild, insulate sequence gives researchers explicit spatial targets for future, region-restricted therapies.
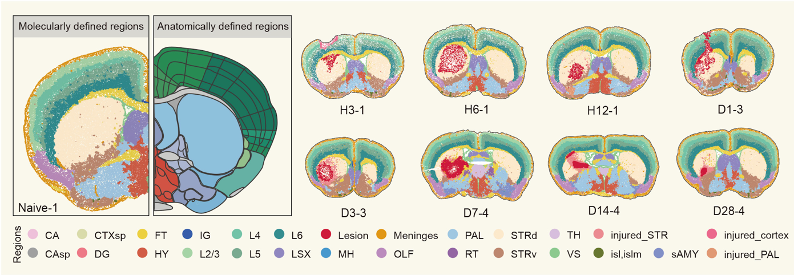 Brain tissue regions defined by gene expression patterns: normal brain anatomy (left) compared to injury-specific regions that emerge after hemorrhage (right).
Brain tissue regions defined by gene expression patterns: normal brain anatomy (left) compared to injury-specific regions that emerge after hemorrhage (right).
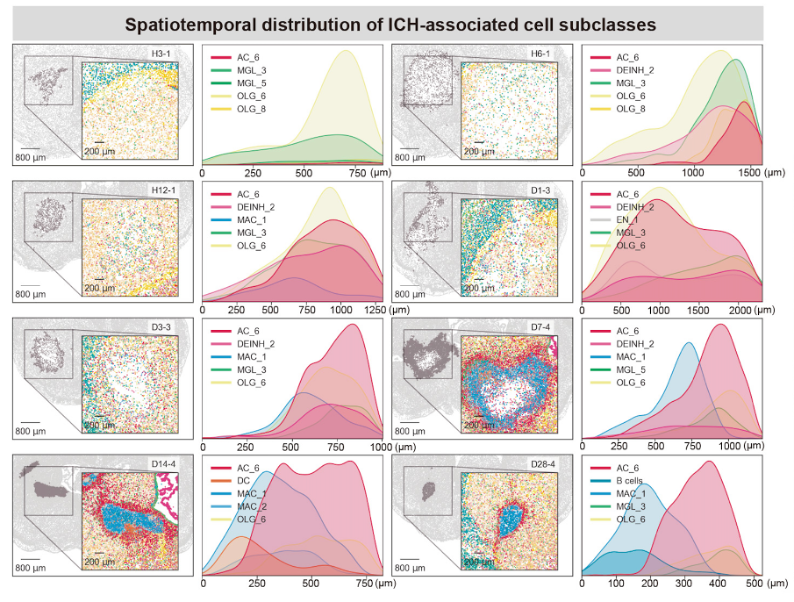 Key cell types redistribute around the lesion over time: astrocytes, microglia, macrophages and oligodendrocytes move closer to the injury site as healing progresses.
Key cell types redistribute around the lesion over time: astrocytes, microglia, macrophages and oligodendrocytes move closer to the injury site as healing progresses.
In spatial transcriptomic research, time is as critical as location. By layering consecutive slices, the authors reconstruct a choreography beginning with chemokine bursts attracting peripheral immune cells within six hours, progressing through a surge of cell-cycle genes and phagocytic activity between day three and seven, and culminating in adaptive immunity, collagen deposition, and immunoglobulin production between the second and fourth weeks. Specific gene modules exhibit peak expression precisely within these windows, providing a molecular time code exploitable by drug developers. A treatment that blocks excessive lipid loading in macrophages, for instance, will be most effective if delivered just before the "response to lipoprotein particle" module ignites.
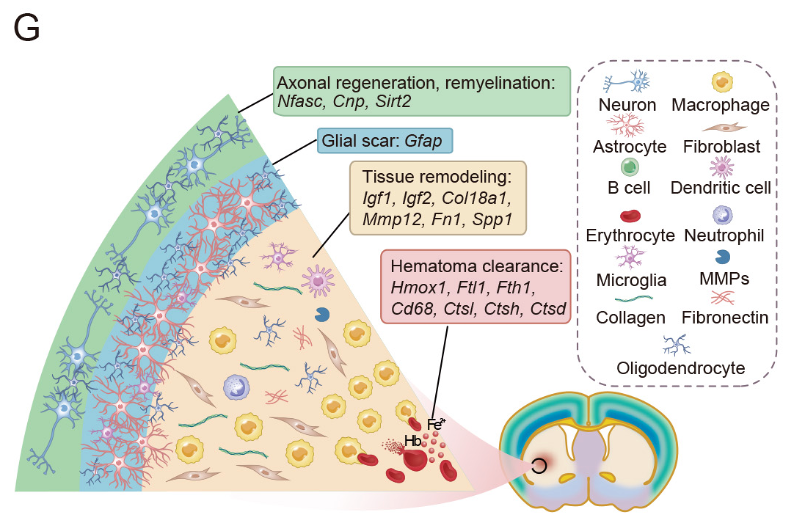 Four distinct zones emerge around the lesion: hemoglobin breakdown at the core, matrix remodeling in the middle layers, astrocytic scarring, and neural repair genes at the periphery.
Four distinct zones emerge around the lesion: hemoglobin breakdown at the core, matrix remodeling in the middle layers, astrocytic scarring, and neural repair genes at the periphery.
The authors emphasize that the mouse findings align with human pathology: astrocytic markers PTX3 and TGM1, shown here to surge in the acute phase, are similarly elevated in perihematomal tissue from patients who die within the first week. More broadly, the open atlas serves as a searchable framework for clinicians and basic scientists to build imaging biomarkers, test neuroprotective compounds or design biomaterial scaffolds that match the brain's own repair timeline. The study demonstrates how spatial transcriptomics can illuminate the complex cellular responses and molecular signaling pathways that determine ICH prognosis. For decades, brain hemorrhage has been treated as an unpredictable catastrophe; now, scientists can observe its progression at key time points, offering opportunities for timely and targeted interventions.
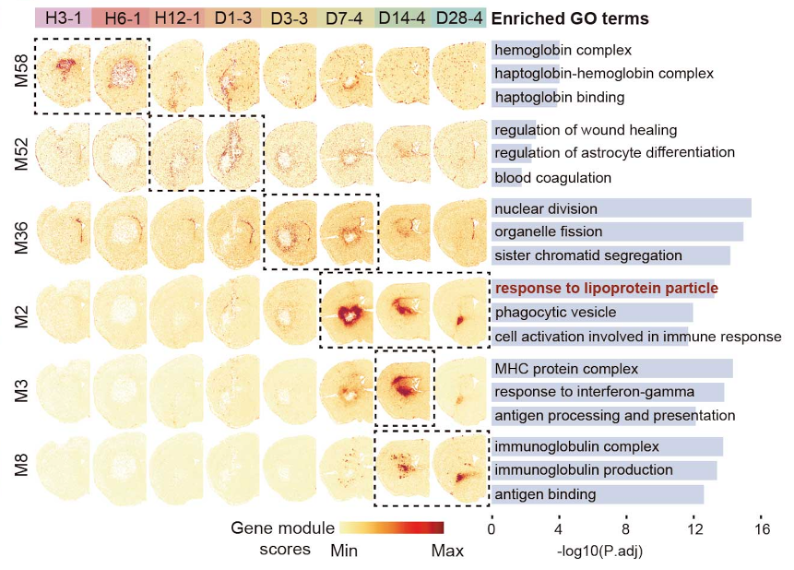 Molecular waves sweep through the injured brain: early hemoglobin response and wound healing give way to proliferation, lipid handling, and late-stage immune responses.
Molecular waves sweep through the injured brain: early hemoglobin response and wound healing give way to proliferation, lipid handling, and late-stage immune responses.
This study enhances the understanding of the local and global brain cell responses to ICH and provides a valuable resource for advancing future research and developing novel therapies for this devastating condition. The findings suggest that targeting specific cell types and molecular pathways at precise time windows could provide new approaches for treating intracerebral hemorrhage patients.
The study's integrated design, spanning single-cell discovery across multiple time points and validation with human tissue, offers a blueprint for understanding sterile inflammatory injury well beyond stroke. Critically important for advancing precision medicine in neuroscience, this breakthrough showcases the transformative power of BGI's spatial omics ecosystem: Stereo-seq's unparalleled subcellular resolution enabled the capture of transcripts from more than 3.3 million cells while preserving their precise tissue architecture, a feat unachievable with conventional single-cell technologies. The platform's ability to analyze formaldehyde-fixed brain tissue sections with exquisite spatial fidelity has opened new frontiers for studying complex neurological diseases.
Ethical approval for this study was obtained. All Stereo-seq data can be explored at https://db.cngb.org/stomics/stmich/ (accession STT0000047). This research can be assessed here: https://doi.org/10.1016/j.neuron.2025.04.026



