Ants march across almost every terrestrial habitat, from scorching dunes to rainforest canopies, yet their real triumph lies in the way each colony functions as a tightly cho-reographed "superorganism" whose sterile workers labour for a single reproductive queen. What underpins their extraordinary success? How did ant societies evolve? And what genomic changes allowed that leap from solitary insect to social empire and helped generate more than 15,000 extant species?
On June 16, an international consortium co-organised by BGI-Research, the Global Ant Genomics Alliance (GAGA), delivers the most comprehensive answer to date. By comparing 163 high-quality ant genomes drawn from 12 of the 16 ant subfamilies, in-cluding 145 newly sequenced species, the team reconstructs the tree of life for ants and dates the origin of modern ants to the late Jurassic about 157 million years ago, uncovers a burst of smell-related gene expansions in the ant ancestor, shows that the rate at which chromosomes rearrange predicts species richness, and identifies deeply conserved gene-regulatory networks, centred on juvenile hormone, insulin and MAPK signalling, that have been repeatedly rewired to sculpt queens, workers and soldiers. The work, published in Cell, not only redraws key branches of ant phylogeny but also lays a blueprint for studying social complexity across the tree of life.
 The study “Adaptive radiation and social evolution of the ants” was published in Cell.
The study “Adaptive radiation and social evolution of the ants” was published in Cell.
Those high-quality ant genomes exposed an unexpected pattern: lineages with the most drastic genome rearrangement (measured by how frequently chromosomes break and rearrange) also exhibit the richest species counts, a correlation that holds across ten subfamilies and exceeds vertebrate rearrangement rates by up to 4.5-fold. Yet within that turbulence the researchers found islands of stability: 970 conserved gene clusters, comprising roughly 3,700 genes, that maintain their synteny in more than 80% of species and display co-expression (implying they work together) in caste development. One such cluster harbours egg production genes flanked by fat metabo-lism genes that activate in future queens, while another bundles hormone and growth regulators that switch on in worker larvae, an elegant example of ancient gene neigh-bourhoods being reused to orchestrate modern social roles.
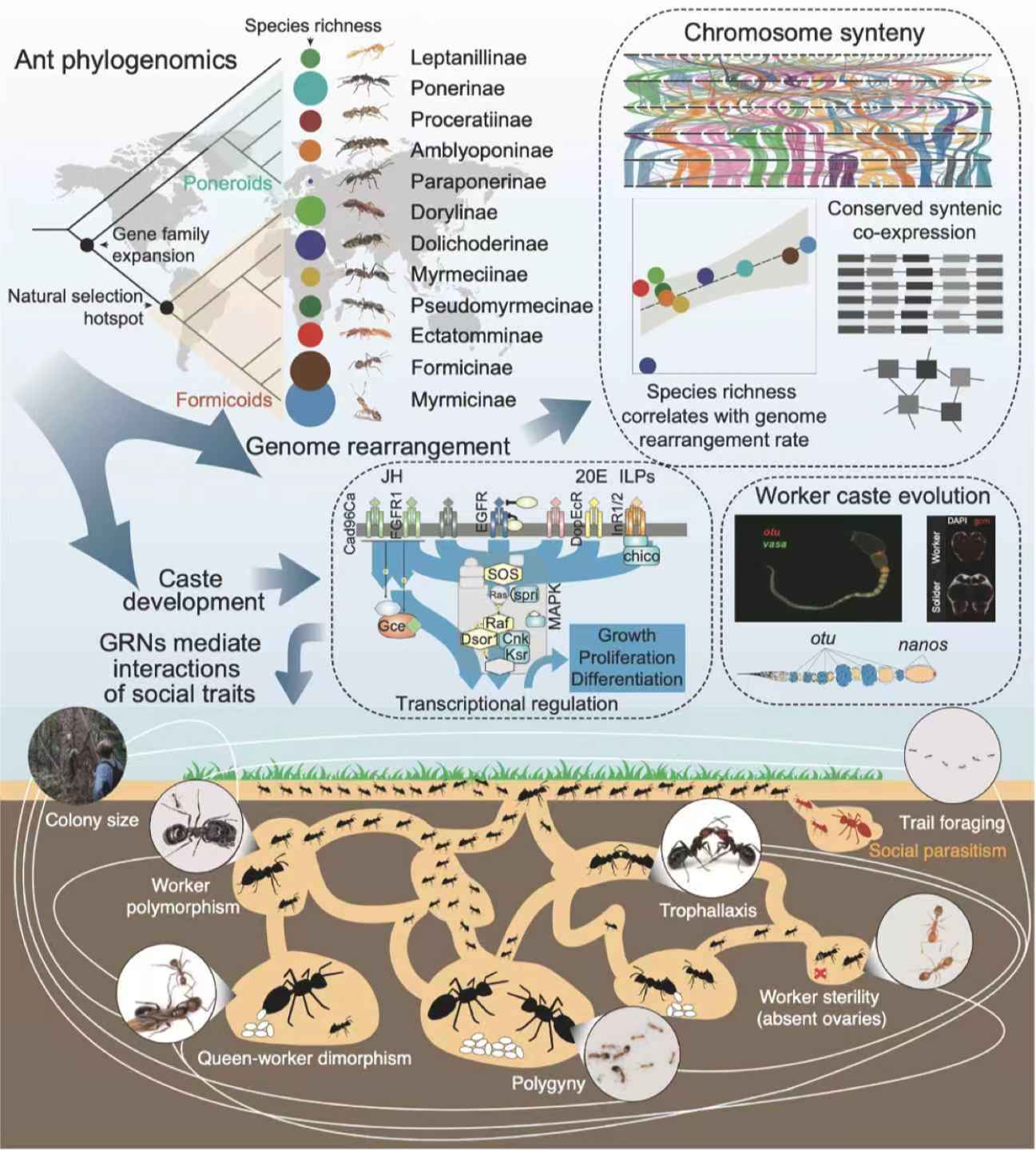 Graphical abstract of the study.
Graphical abstract of the study.
Natural selection, the study shows, has repeatedly targeted signaling pathways famil-iar from developmental biology but pressed into new social service. In the common ancestor of the hyper-diverse formicoid ants, today accounting for 90% of extant spe-cies, hormone processing enzymes and detoxification gene families expanded dra-matically, while more than 200 single-copy genes involved in DNA organization, im-munity and longevity show strong signatures of evolutionary pressure. Later elabora-tions, such as the evolution of wingless "ergatoid" queens or worker driven reproduc-tion in gamergate species, left distinct footprints of intensified or relaxed selection on MAPK and insulin receptors, underscoring how a core toolkit can be dialled up or down to fit contrasting lifestyles. Pharmacological proof came from Monomorium phar-aonis: blocking MAPK with trametinib, a specific inhibitor drug, swelled worker body length, blurring caste boundaries and validating the pathway's developmental role.
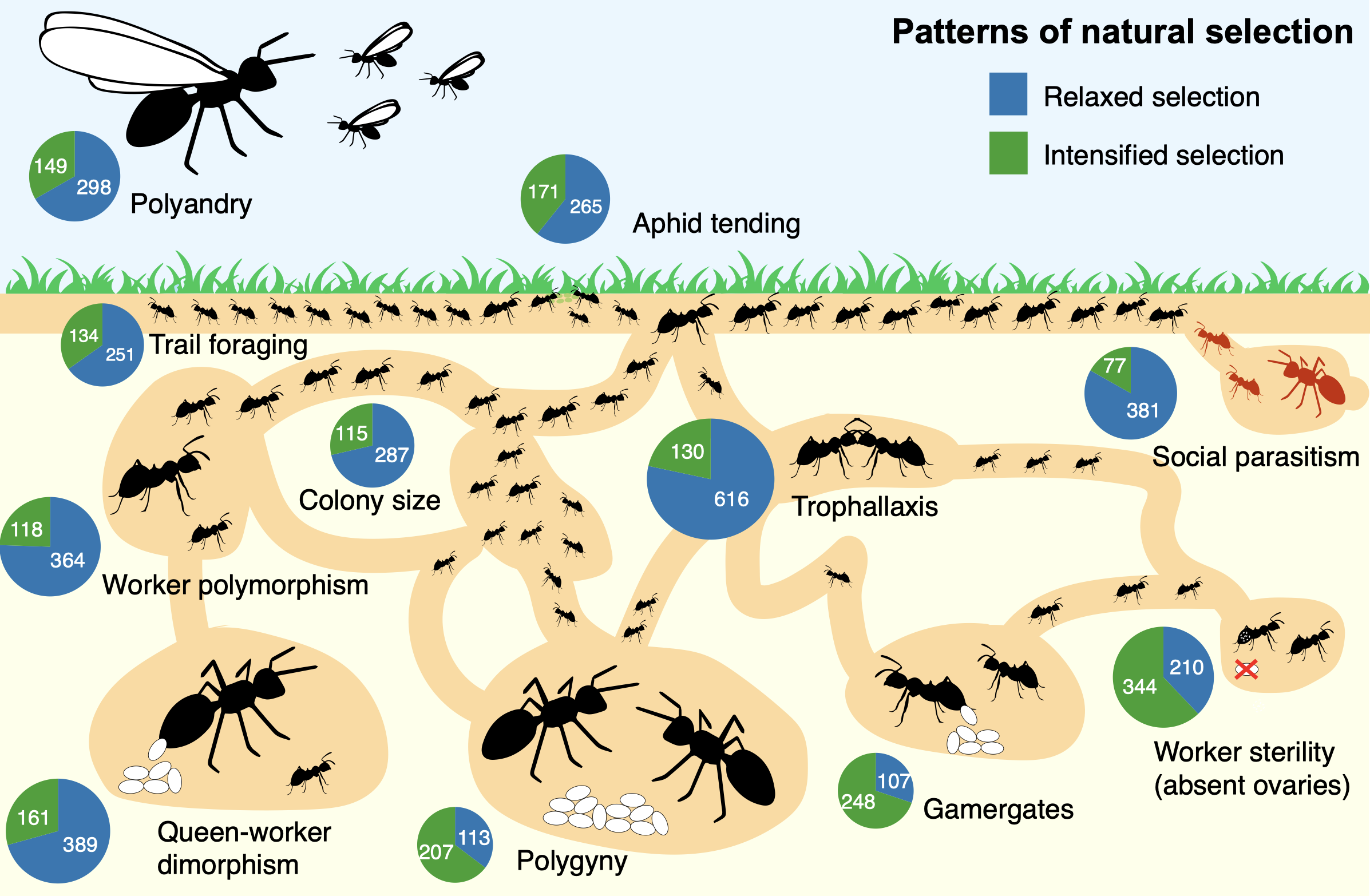 Eleven social traits that evolved repeatedly while leaving consistent signatures of in-tensified (green) and relaxed (blue) selection (gene numbers in pies). (Picture by Joel Vizueta)
Eleven social traits that evolved repeatedly while leaving consistent signatures of in-tensified (green) and relaxed (blue) selection (gene numbers in pies). (Picture by Joel Vizueta)
As noted by the research team, this genome atlas reveals the evolutionary levers that transformed small, solitary wasp-like ancestors into the world's dominant engineers of soil and canopy. The integrated trait matrix finally connects specific genes to complex social phenotypes, providing a framework for dissecting division of labour in any eu-social animal, as described in the study. Looking ahead, the research establishes foundations for future comprehensive molecular investigations into queen longevity, ant fungus farming and the biological costs of chromosomal rearrangement.
All raw reads, assemblies and annotation files have been deposited in the NCBI As-sembly and Sequence Read Archive under BioProjects PRJNA1172379 and PRJNA1159026, with analysis code available on GitHub. The consortium invites evo-lutionary biologists, ecologists and synthetic biology engineers to mine the dataset and explore how conserved molecular circuits can be rewired to build cooperative sys-tems, whether they walk on six legs or sit on a silicon wafer.
This research can be assessed here: https://doi.org/10.1016/j.cell.2025.05.030



