Yunnan University and BGI-Research have successfully constructed a reference set of 14,062 gut microbiota from animals living on the Qinghai-Tibet Plateau, revealing rich microbial resources. The results were reported online on November 20, 2025, in the scientific journal Microbiome in a paper titled “A Unified Catalog of 14,062 Microbial Species Reference Genomes Provides New Insight into the Gut Microbiota in High Altitude Mammals”.
The study “A Unified Catalog of 14,062 Microbial Species Reference Genomes Provides New Insight into the Gut Microbiota in High Altitude Mammals” was published in Microbiome.
Genomic data from the study has been made publicly available, crucially supporting the potential development of novel gene-editing tools, antimicrobial peptides and other biotechnology products.
With an average elevation of 4,500 meters, the Qinghai-Tibet Plateau, the world’s largest and highest plateau above sea level, is a unique natural laboratory for studying life’s adaptations to extreme environments.
To construct the gut microbiota database the scientific research team from Yunnan University and BGI-Research spent five years systematically collecting more than 5,000 fresh fecal samples from six herbivorous mammals that have known phylogenetic relationships and are native to the Qinghai-Tibet Plateau; the Tibetan antelope, Tibetan cattle, Tibetan sheep, Tibetan horse, Tibetan ass, and the yak.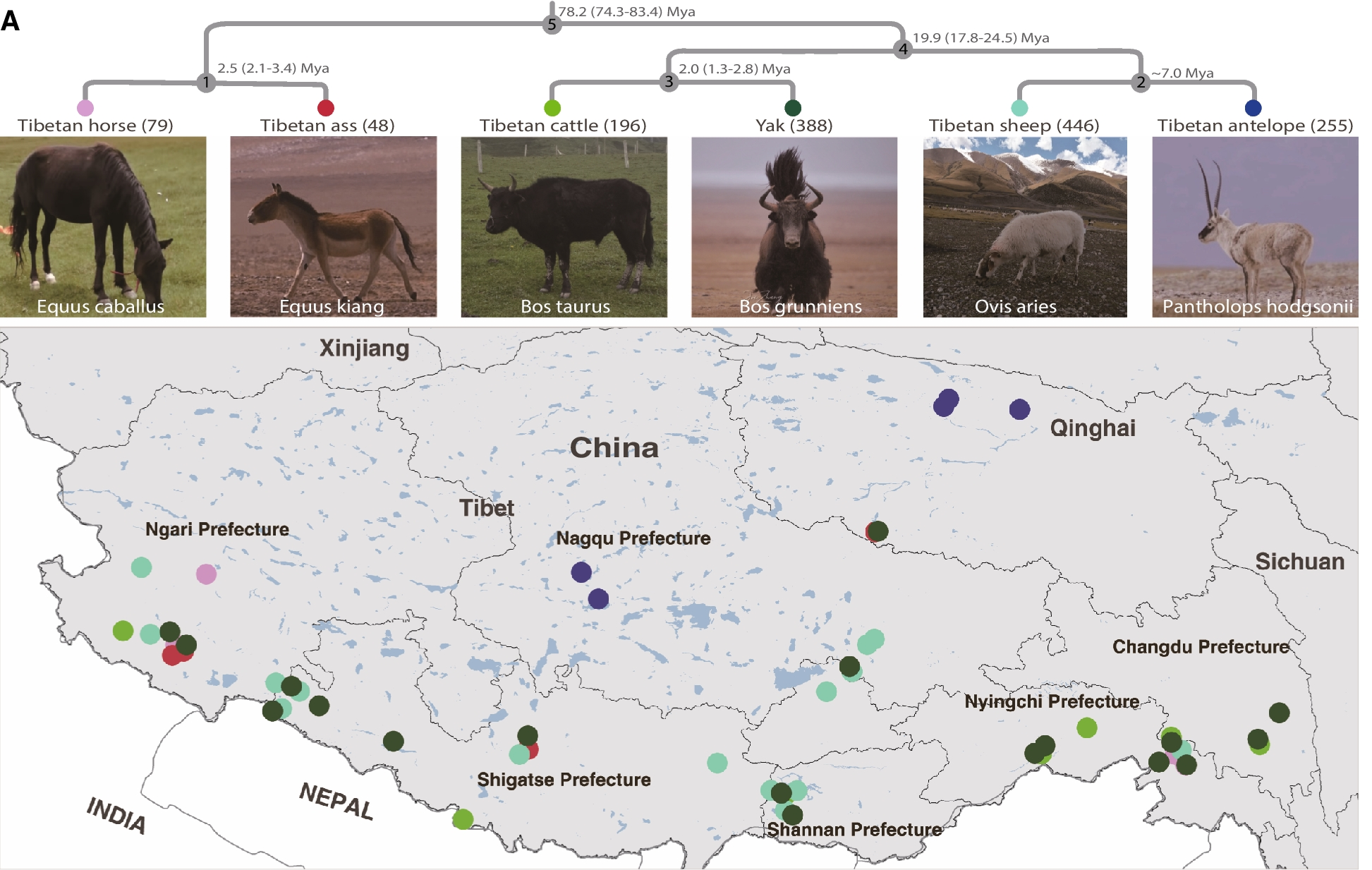
The phylogenetic tree of six host animal species living in the Qinghai-Tibetan Plateau and geographical distribution of collected fecal samples.
The gut microbiota is essential for health and survival and in the phase of the study reported in Microbiome, the inner part of 1,412 fresh fecal samples were collected, recorded and stored at -80oC before being transported to the China National Gene Bank where metagenomic sequencing yielded more than 33Tb of data.
The team then employed two methods of cross-validation to successfully construct a high-quality microbial genome reference database containing 14,062 high-confidence species-level genome bins (SBGs). Remarkably, 88% of the microbial species included in the database are previously unknown species, highlighting the microbial resources hidden in the gut of these animals.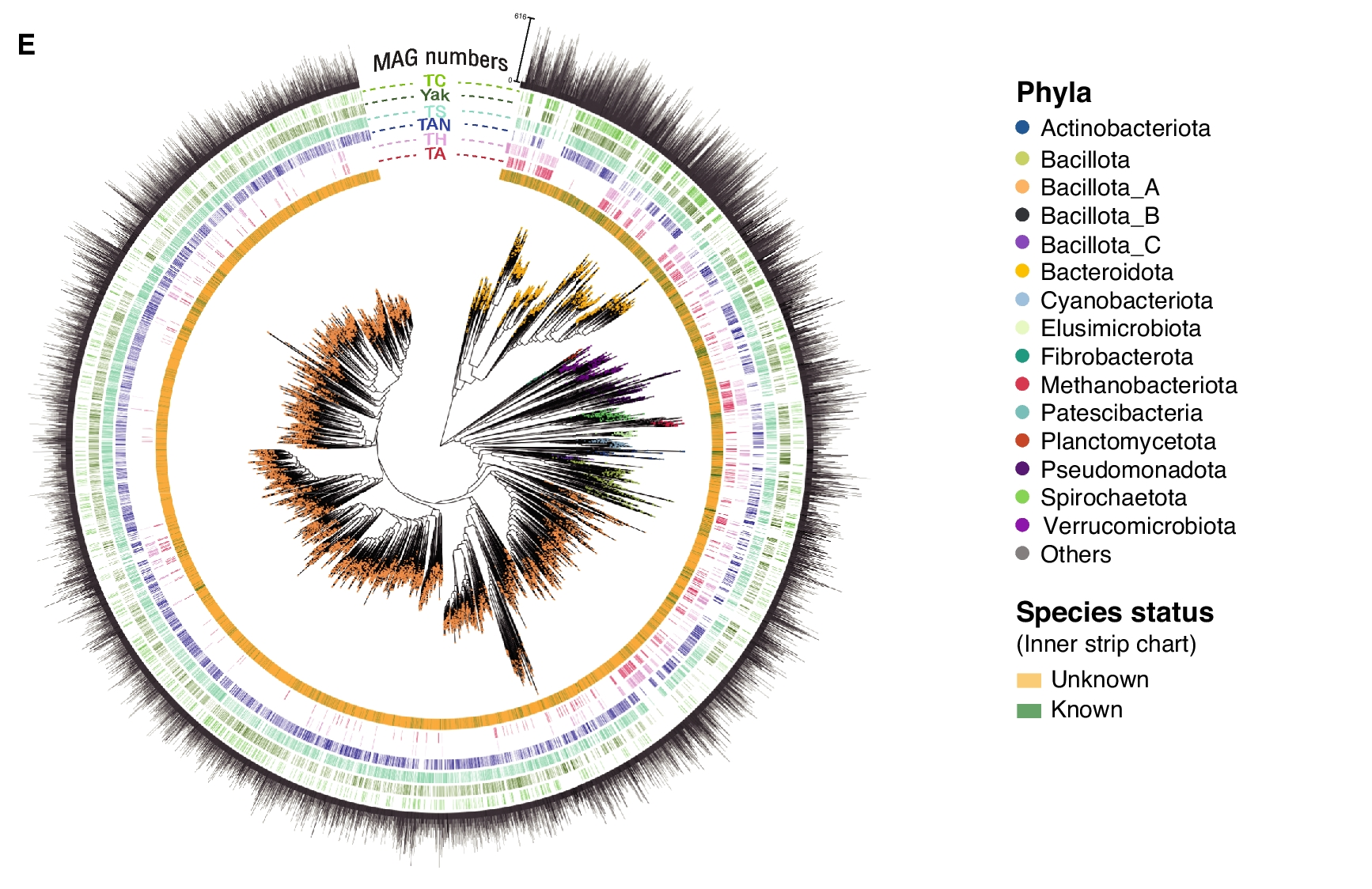
Phylogenetic tree of 14,062 high-confidence species-level genome bins.
Through in-depth mining of this genome database, the research team also discovered that these previously unknown gut microbiota contain a vast number of potentially valuable biosynthetic gene clusters, which offer substantial exploratory value in fields such as biotechnology and drug development.
Li Xiaoping from BGI-Research and co-first author of the paper notes that, “this research project has made us realize that the gut microbiota of animals, especially those in special habitats and with specific diets, is a treasure trove of microbial resources yet to be fully explored, and its further development has profound scientific value and application significance.”
Researcher Zhang Zhigang from Yunnan University and corresponding author of the paper stated that the work not only enriches scientific understanding of the diversity and function of the gut microbiota of large herbivores on the Qinghai-Tibet Plateau, but also provides an empirical basis for understanding the diversity trajectory of host-microbe interactions on an evolutionary scale, which has important reference value for ecology, evolutionary biology, and the development of microbial resources.
At the project's inception, the research team simultaneously conducted pure culture studies, screening nearly 3,000 single bacterial strains from the guts of animals native to the Qinghai-Tibet Plateau. Genomic analysis of over 500 strains from yaks revealed that the gut microbiota of these animals contains a vast and highly diverse array of previously unknown microorganisms, with a potential for novel species accounting for approximately 38%. Furthermore, their genomes were found to possess rich and highly novel secondary metabolic potential.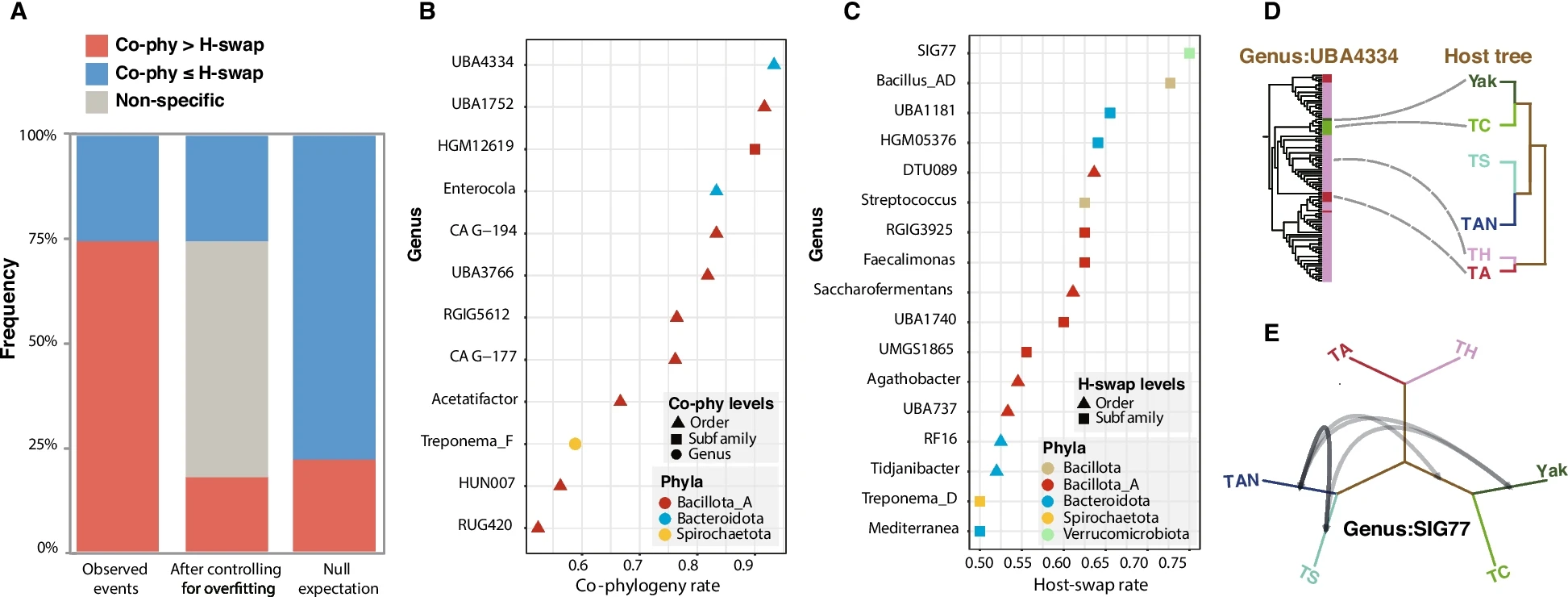
Vertical and horizontal inheritance of high-altitude mammalian gut symbionts.
Biosynthetic gene cluster analysis revealed that over 94% of the predicted gene clusters had no homologs in existing databases, indicating the existence of a large number of unknown natural products. The research team has identified several functional bacterial strains, including 13 novel strains capable of efficiently degrading cellulose and strains with the potential to reduce methane emissions; related in vitro validation experiments are underway. Initial results of this research were published earlier this year in the journal mSystems.
These studies expand BGI Group’s achievements in exploring plateau environmental adaptation, including analysis and sampling at the summit of Mount Everest, adding to life science research in extreme environments.
This study can be accessed here:
https://microbiomejournal.biomedcentral.com/articles/10.1186/s40168-025-02232-5



