On November 20, scientists from BGI-Research, in collaboration with the Yellow Sea Fisheries Research Institute of the Chinese Academy of Fishery Sciences and the Hong Kong University of Science and Technology, report new hologenomic insights into how a deep-sea black coral survives in permanent darkness, low temperature, high pressure and nutrient-poor conditions. Their study, published in Cell Host & Microbe, reveals that this coral hosts a streamlined but highly efficient symbiont community that provides nutritional complementation, waste detoxification, antioxidant support and viral defense.
The study “Hologenomic insights into the molecular adaptation of deep-sea coral Bathypathes pseudoalternata” was published in Cell Host & Microbe.
Deep-sea Coral Bathypathes pseudoalternata.
This study proposes a new paradigm of "simplified, stable and highly complementary" deep-sea symbiosis, offering a molecular framework for understanding how deep-sea coral ecosystems persist in extreme environments. Focusing on the species Bathypathes pseudoalternata (B. pseudoalternata), which forms dense forests on seamount chains in the central South China Sea and Western Pacific Ocean, the research team combined multi-omics analyses with advanced imaging to examine the coral and its microbial partners as a unified, inteconnected biological system.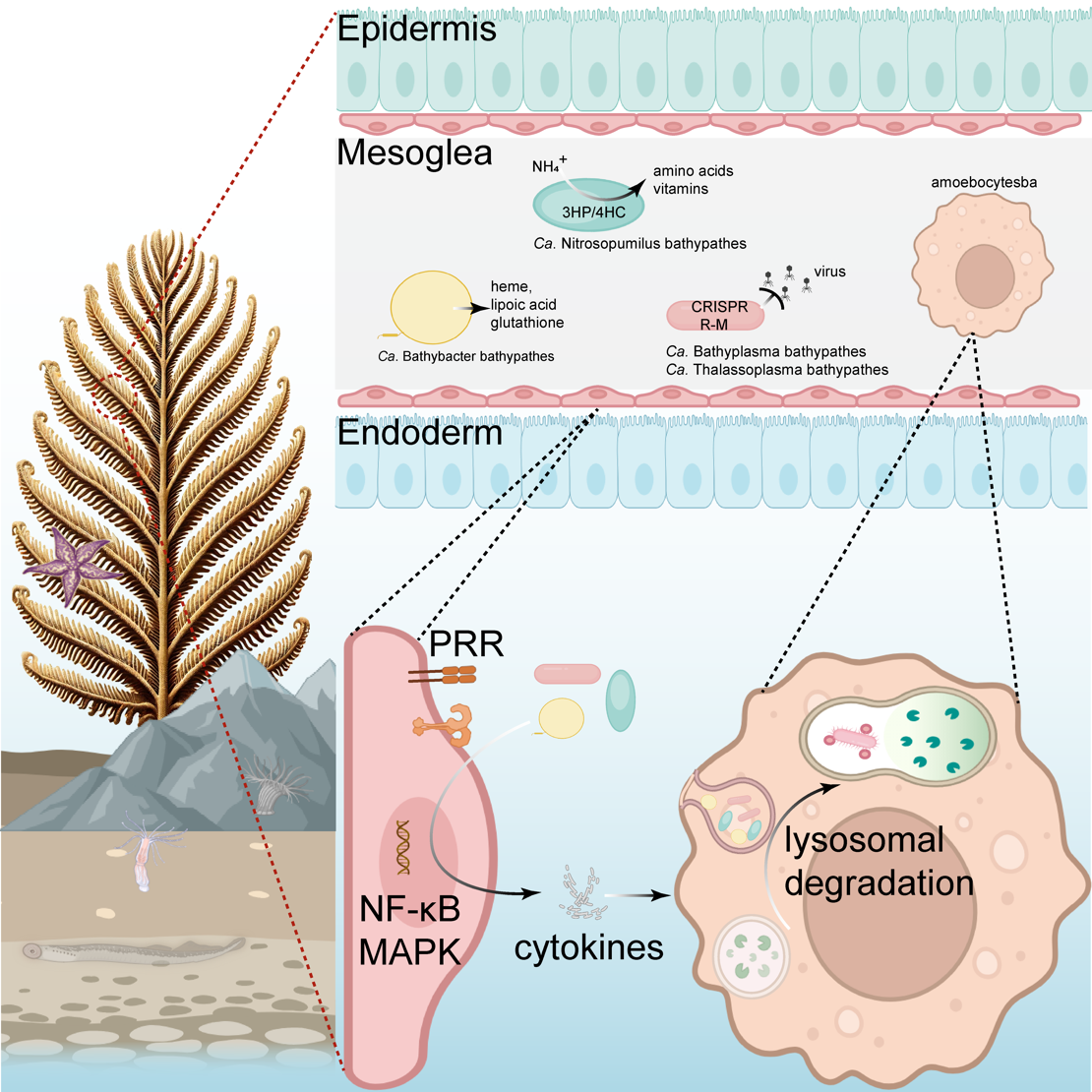
Division of labour in a deep-sea holobiont. The Bathypathes pseudoalternata host and four core microbial partners partition functions for nutrient supply, detoxification, antioxidant production and viral defense, providing a compact blueprint for survival in oligotrophic deep-sea environments.
The researchers have successfully generated a high-quality, chromosome-level genome assembly for B. pseudoalternata spanning approximately 657 megabases across 16 pseudochromosomes, with 29,835 genes and 93.0% completeness. Gene family analyses indicated significant expansions in transporters, immune response genes, and lysosomal enzymes, consistent with an adaptation strategy that reinforces material uptake, immune recognition and controlled intracellular digestion. Notably, the coral genome lacks complete biosynthetic pathways for several amino acids and vitamins, indicating host dependence on symbionts.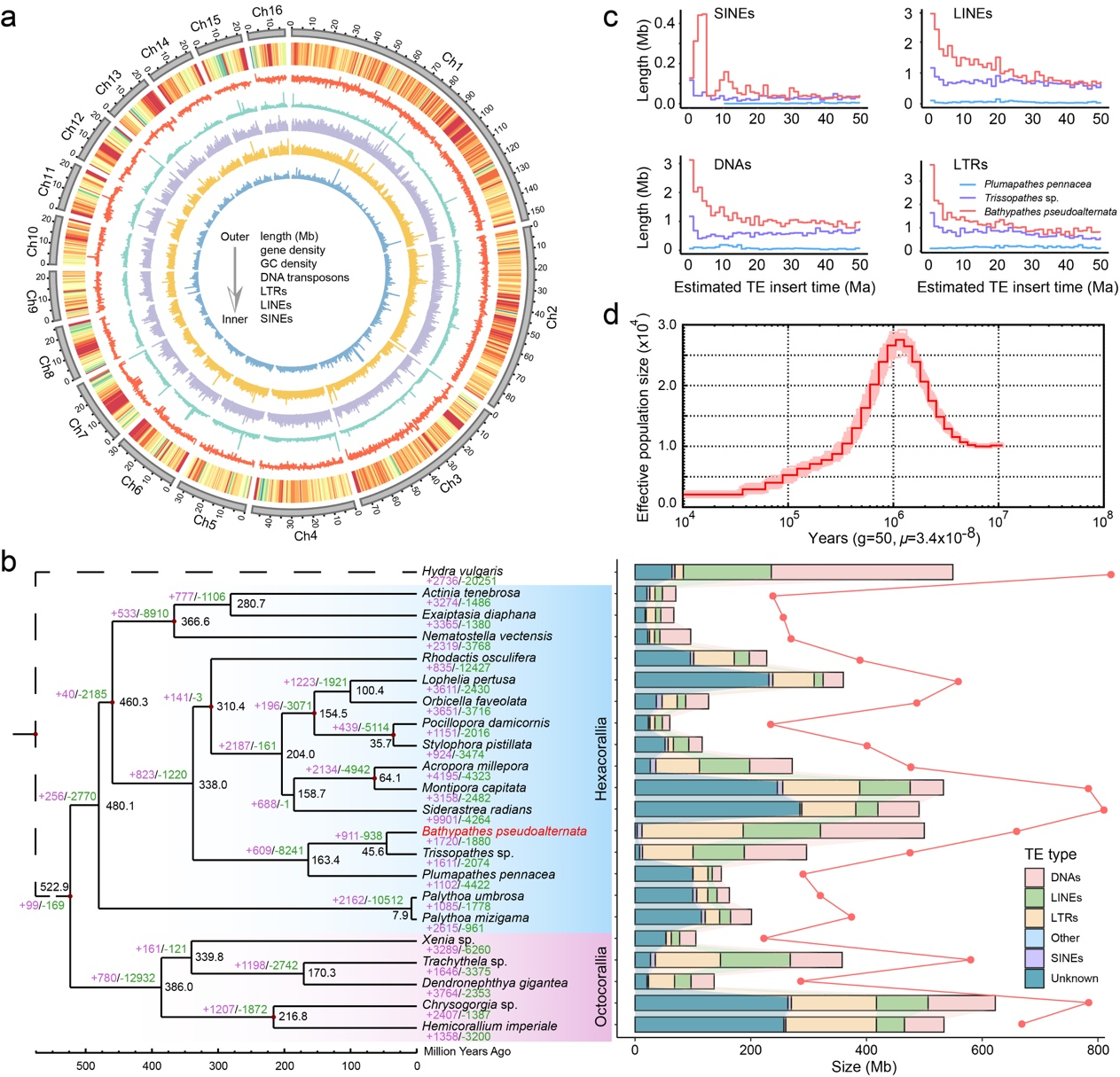
Genome architecture and evolutionary history of B. pseudoalternata. Chromosome-scale assembly, transposable element composition and inferred demographic history reveal how genome expansion and gene family turnover supported the coral's long-term adaptation to deep-sea conditions and stable symbiosis.
The researchers examined microbial communities from 14 coral colonies collected at vatios depths and locations in the South China Sea and Western Pacific Ocean, comparing these to surrounding water and sediment. Amplicon sequencing and metagenomic analyses revealed that the B. pseudoalternata holobiont maintains a remarkably simplified yet stable microbiome dominated by a few closely related lineages, clearly distinct from the more diverse communities in the ambient environment. The dominant symbionts were nearly absent in nearby seawater and sediments, suggesting a specialized, host-specific association. Hybridization chain reaction (HCR) imaging and transmission electron microscopy (TEM) localized these microbes throughout the polyps with strong enrichment in the mesoglea, a collagen-rich, gel-like layer that serves as a shared microenvironment for nutrient exchange and immune modulation.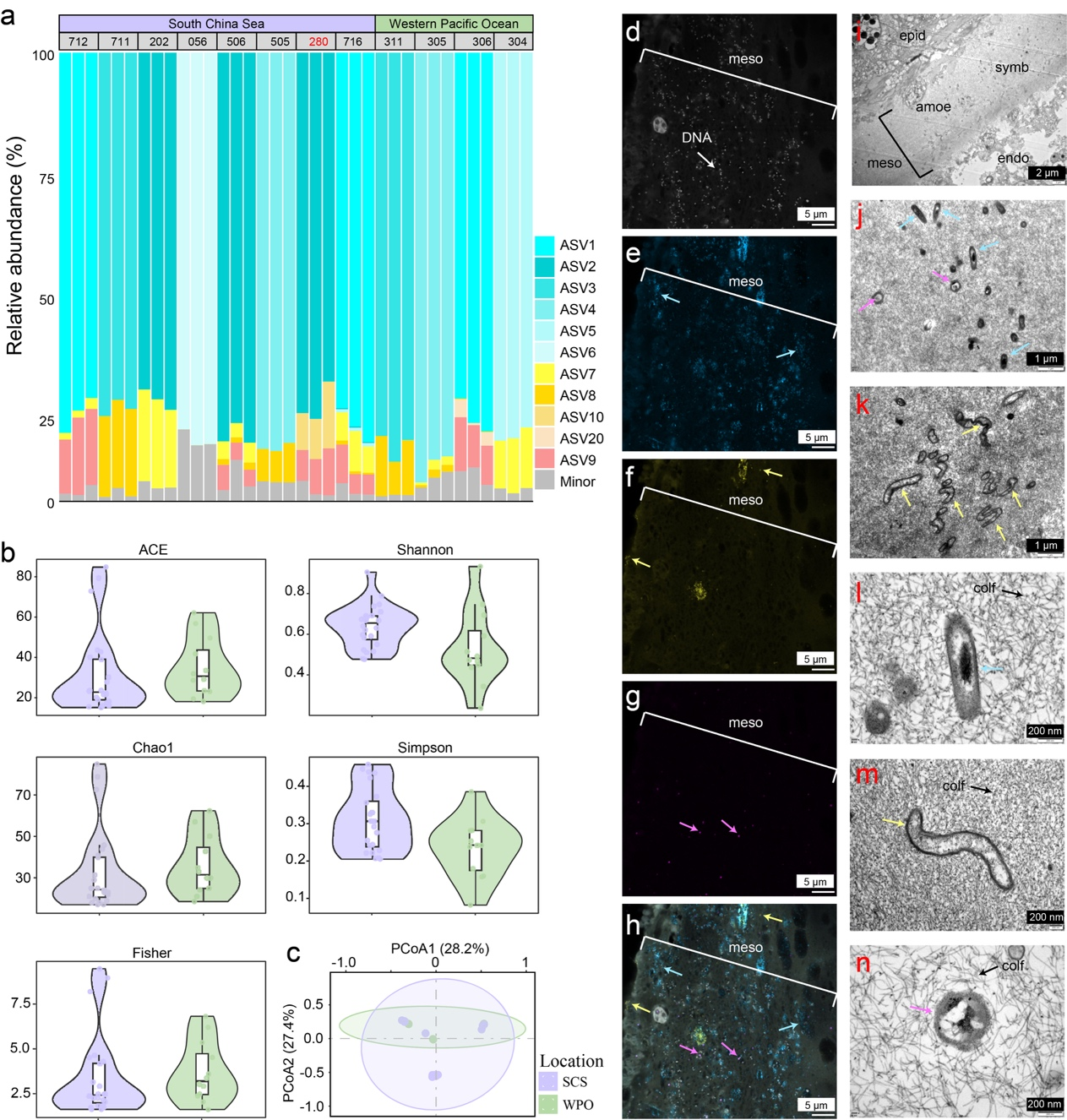
Microbial community structure and tissue localization in the B. pseudoalternata holobiont. A small number of dominant symbionts form a stable, host-specific consortium concentrated in the mesoglea, supporting a model of streamlined yet spatially organized deep-sea symbiosis.
By combining metagenome-assembled genomes with transcriptomic data, the researchers reconstructed metabolic contributions for four core symbionts. Candidatus Nitrosopumilus bathypathes, the dominant archaeal partner accounting for approximately 78% of the microbiome, oxidizes host-derived ammonia to gain energy for synthesizing multiple amino acids and vitamins that are incomplete or missing in the host biosynthetic repertoire, coupling detoxification with nutrient supply. Ca. Bathybacter bathypathes, a newly identified alphaproteobacterial symbiont representing a novel order and accounting for approximately 8% of the microbiome, appears capable of synthesizing heme, lipoic acid, glutathione and fatty acids, providing both antioxidants and nutrients. Two Mollicutes-like species, Ca. Bathyplasma bathypathes and Ca. Thalassoplasma bathypathes, together accounting for approximately 11% of the microbiome, have highly reduced genomes that retain a Type II CRISPR-Cas system and multiple restriction–modification systems, respectively, forming a microbial shield against viral invasion.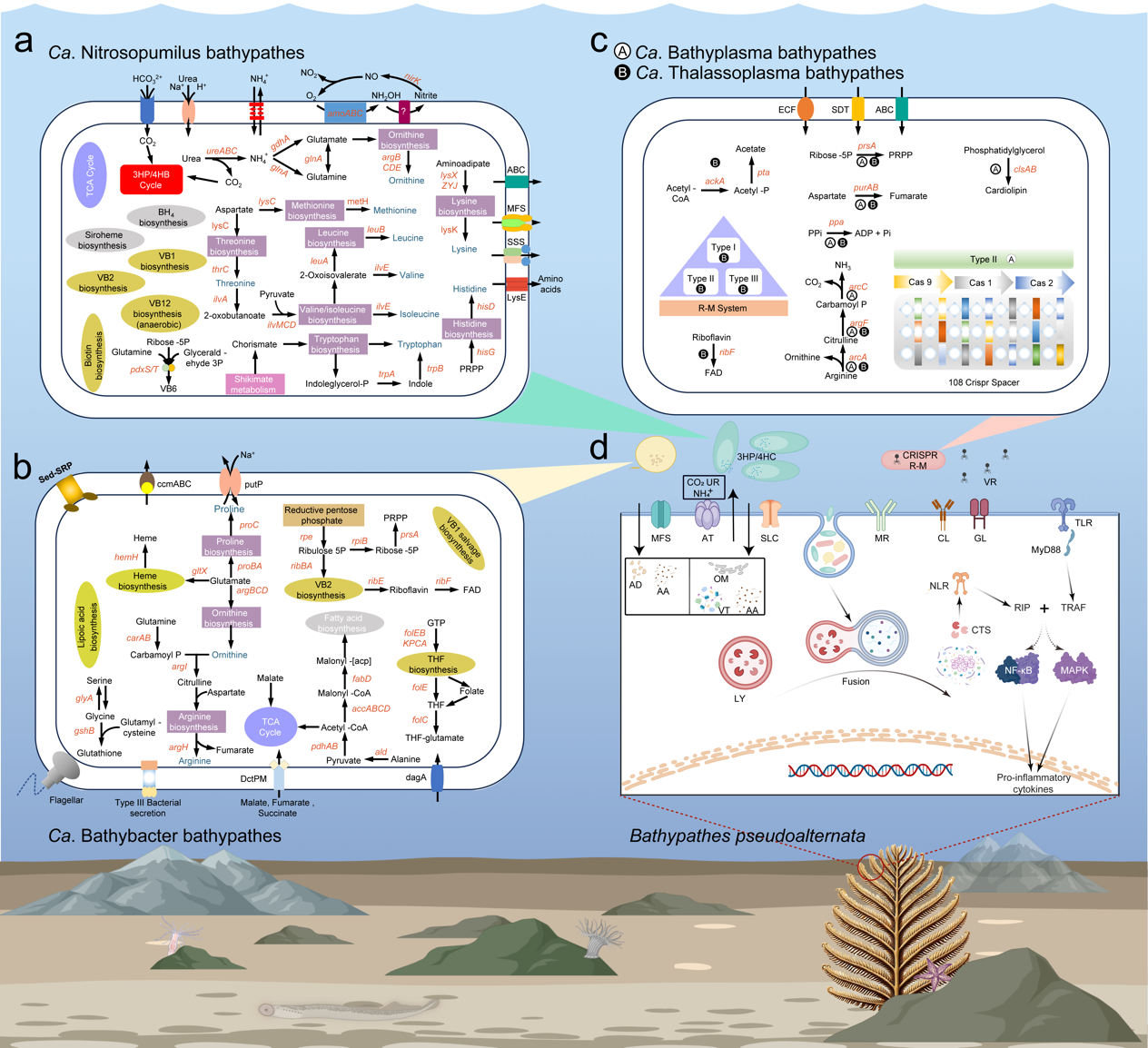
Metabolic pathways and host-symbiont interactions in the B. pseudoalternata holobiont. Four core symbionts execute specialized metabolic functions including nitrogen cycling, antioxidant synthesis, and viral defense, while the host coordinates bidirectional nutrient exchange and immune-mediated symbiont management, forming an integrated network that sustains the holobiont in extreme deep-sea conditions.
On the host side, transcriptional profiling showed high activity of pattern recognition receptors and expanded gene families involved in innate immunity and lysosomal enzymes. The authors suggest that the host uses immune surveillance to detect microbial patterns and recruit immune cells to carry out controlled phagocytosis and lysosomal digestion of symbionts. This balanced process supports the stable coexistence of a select few highly specialized microbial partners, thoughtfully maintaining the coral’s nutritional and defensive requirements.
By revealing the molecular mechanisms that connect nutrient acquisition, ammonia detoxification, antioxidant protection, viral defense and immune homeostasis within a single, spatially organized system, this study provides a valuable framework for understanding how deep-sea corals contribute to biogeochemical cycling and how resilient their ecosystems may be to environmental change. Such holobiont-level insights can support future assessments of deep-sea biodiversity and guide the exploration of deep-sea functional gene resources, while highlighting the importance of conserving these slowly growing habitat-forming species.
All coral sampling and experimental procedures were carried out in accordance with relevant national and institutional guidelines for the collection and study of marine invertebrates, and we gratefully acknowledge the approval and support provided by the appropriate authorities.
This research can be assessed here:
https://www.cell.com/cell-host-microbe/abstract/S1931-3128(25)00454-8



