A recent study led by BGI-Research, titled "Tracing the tissue origin of cell-free DNA through open chromatin footprint," has been published online in the renowned journal Communications Biology.
In this study, researchers introduced a novel approach known as the Tissue Contribution Index (TCI). This innovative technique addresses critical bottlenecks in existing methods for tracing the tissue origin of plasma cell-free DNA (cfDNA)—specifically issues related to DNA damage, high costs, and limited clinical applicability. By leveraging open chromatin footprints, TCI provides a more efficient and precise tool for diverse clinical scenarios, including non-invasive prenatal testing (NIPT), organ transplant monitoring, cancer diagnosis, and prognosis assessment for infectious diseases.
 The study “Tracing the tissue origin of cell-free DNA through open chromatin footprint” was published open access in Communications Biology (Published 25 November 2025; DOI: https://doi.org/10.1038/s42003-025-09232-z).
The study “Tracing the tissue origin of cell-free DNA through open chromatin footprint” was published open access in Communications Biology (Published 25 November 2025; DOI: https://doi.org/10.1038/s42003-025-09232-z).
Plasma cfDNA is a promising biomarker for liquid biopsy, essential for diagnosing and monitoring diseases. However, traditional methods for estimating tissue contributions—primarily relying on methylation markers—have long faced significant limitations. These approaches often involve complex processing, such as bisulfite sequencing, that can damage DNA samples. They also tend to incur high costs and struggle to provide precise quantification, thereby limiting their widespread clinical adoption.
To address these challenges, the research team developed TCI by leveraging a distinct biological phenomenon: the depletion of cfDNA coverage near the transcriptional start site (TSS) of actively transcribed genes due to open chromatin. By utilizing these fragmentation patterns in tissue-specific highly expressed genes (SH-genes), TCI achieves precise tracking of tissue origins without the need for complex pre-processing.
This approach not only preserves sample integrity but is also compatible with various sequencing modes (including single-end sequencing), significantly reducing detection costs. Crucially, TCI allows for the clear quantification of tissue contributions to plasma cfDNA, effectively overcoming the fundamental limitations of traditional methodologies.
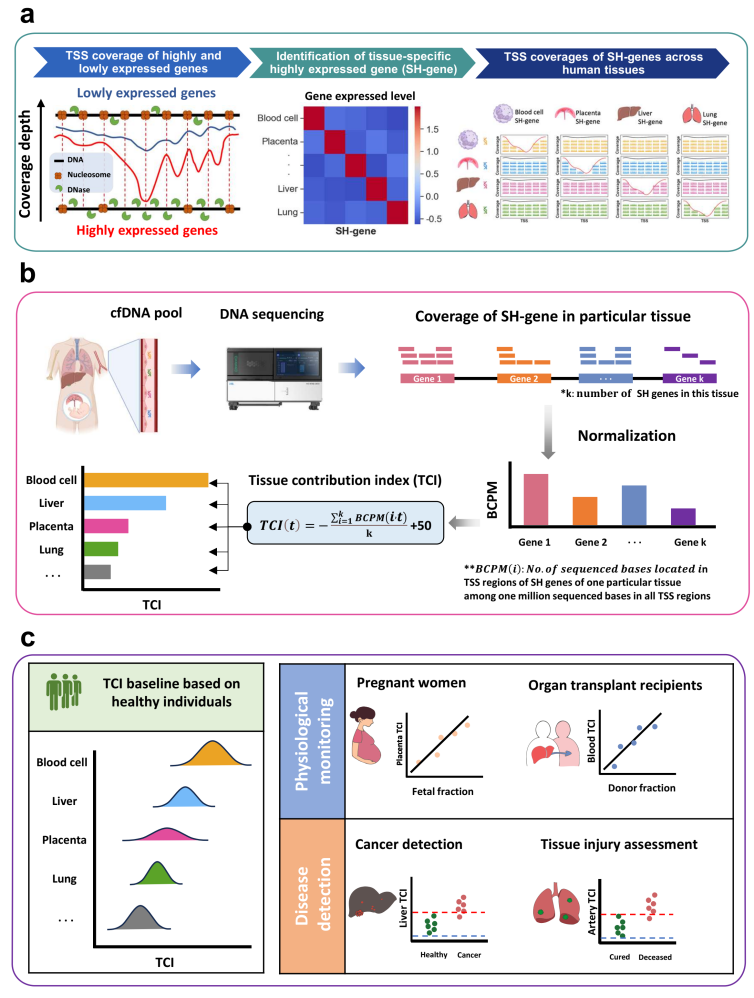
Study Overview of the Tissue Contribution Index (TCI).
The research team rigorously validated TCI across diverse potential clinical application scenarios, proving its robustness in both physiological monitoring and disease detection.
In the realm of monitoring, the method demonstrated exceptional precision. For prenatal care, TCI accurately tracked fetal DNA fractions by analyzing placental signals, offering a streamlined alternative to NIPT that eliminates the need for specific genetic markers like SNPs. Similarly, in organ transplant recipients, the technology effectively traced dynamic changes in donor-derived cfDNA. This capability allows clinicians to closely monitor donor organ health and timely detect potential risks, such as tissue damage or immune rejection events like acute Graft-Versus-Host Disease.
Beyond monitoring, TCI showed significant promise in diagnostics and prognosis. When applied to infectious diseases—specifically validated using data from COVID-19 patients—the tool successfully evaluated tissue damage in the lungs and arteries to predict disease progression and support risk stratification. Crucially, the study also highlighted TCI’s potential in oncology. In tests involving Hepatocellular Carcinoma (HCC), the "Liver TCI" successfully identified abnormal DNA signals from tumor tissues. The results showed that TCI achieved high diagnostic performance, outperforming traditional fragmentomic biomarkers such as fragment size and end motifs.
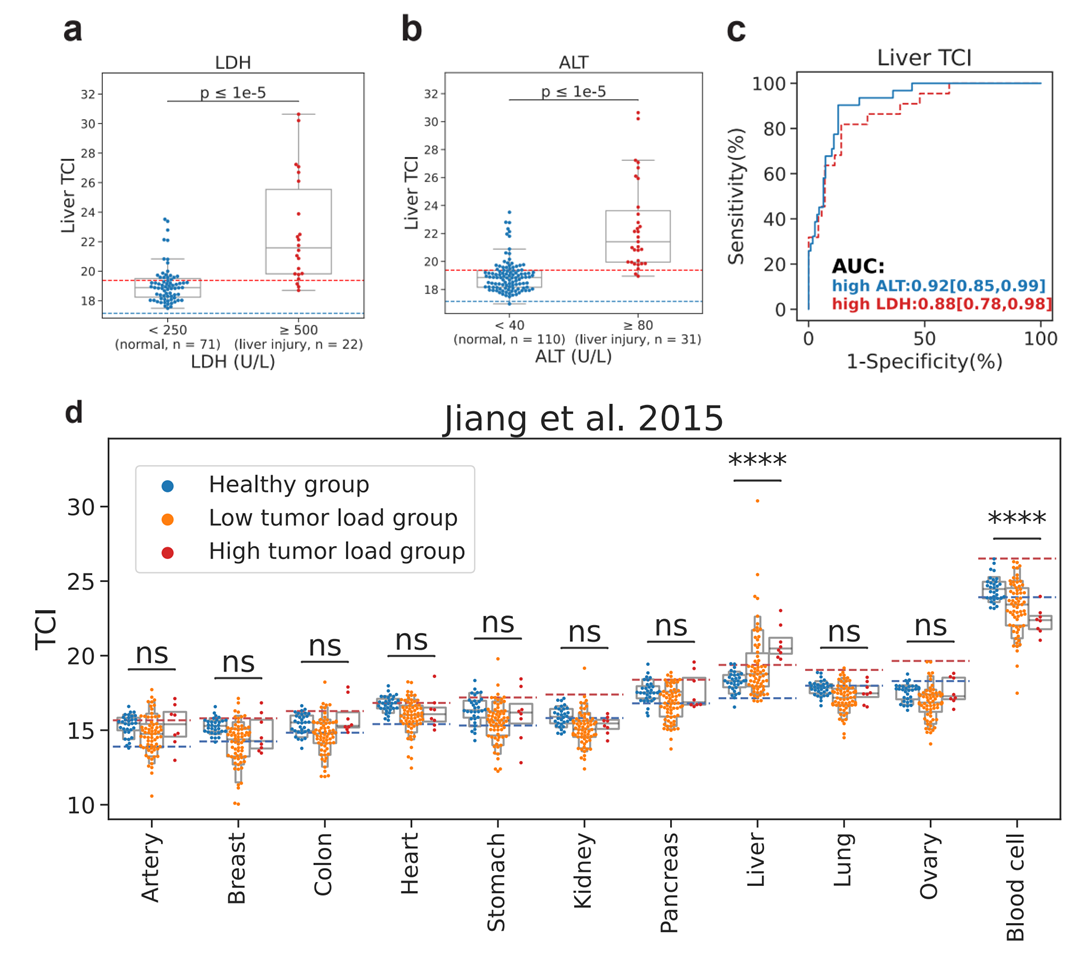 Clinical Application of TCI in Liver Disease Detection.
Clinical Application of TCI in Liver Disease Detection.
Furthermore, utilizing plasma DNA samples from 460 healthy individuals, the study established "reference intervals" for TCI across different tissues. This defines the normal range of TCI under healthy conditions. These reference intervals serve as a critical benchmark for identifying abnormal tissue contributions, significantly enhancing the clinical value of TCI for detecting disease-related anomalies.
"The key innovation of TCI is that it combines precise quantification of cfDNA tissue origin with clinical accessibility," said Dr. Haiqiang Zhang, a researcher at BGI-Research who co-led the study with Dr. Xin Jin. "Traditional methods either damage cfDNA molecules or are too costly for widespread use. In contrast, TCI is compatible with cost-effective single-end sequencing. It allows for the simultaneous analysis of tissue contribution and fragmentomic characteristics, providing 'one-stop' information for clinicians."
Dr. Zhang added, "In the future, we plan to further identify tumor-specific genes to enhance sensitivity for early cancer screening and establish TCI reference intervals for diverse populations to better meet clinical needs."
The sequencing data from this study have been deposited in the China National GeneBank Sequence Archive (CNSA). To facilitate further verification and application by global research and clinical institutions, the implementation code for the TCI method has been made open-source and is available on GitHub: https://github.com/lingguoli/TCI.
This research is available at: https://www.nature.com/articles/s42003-025-09232-z



