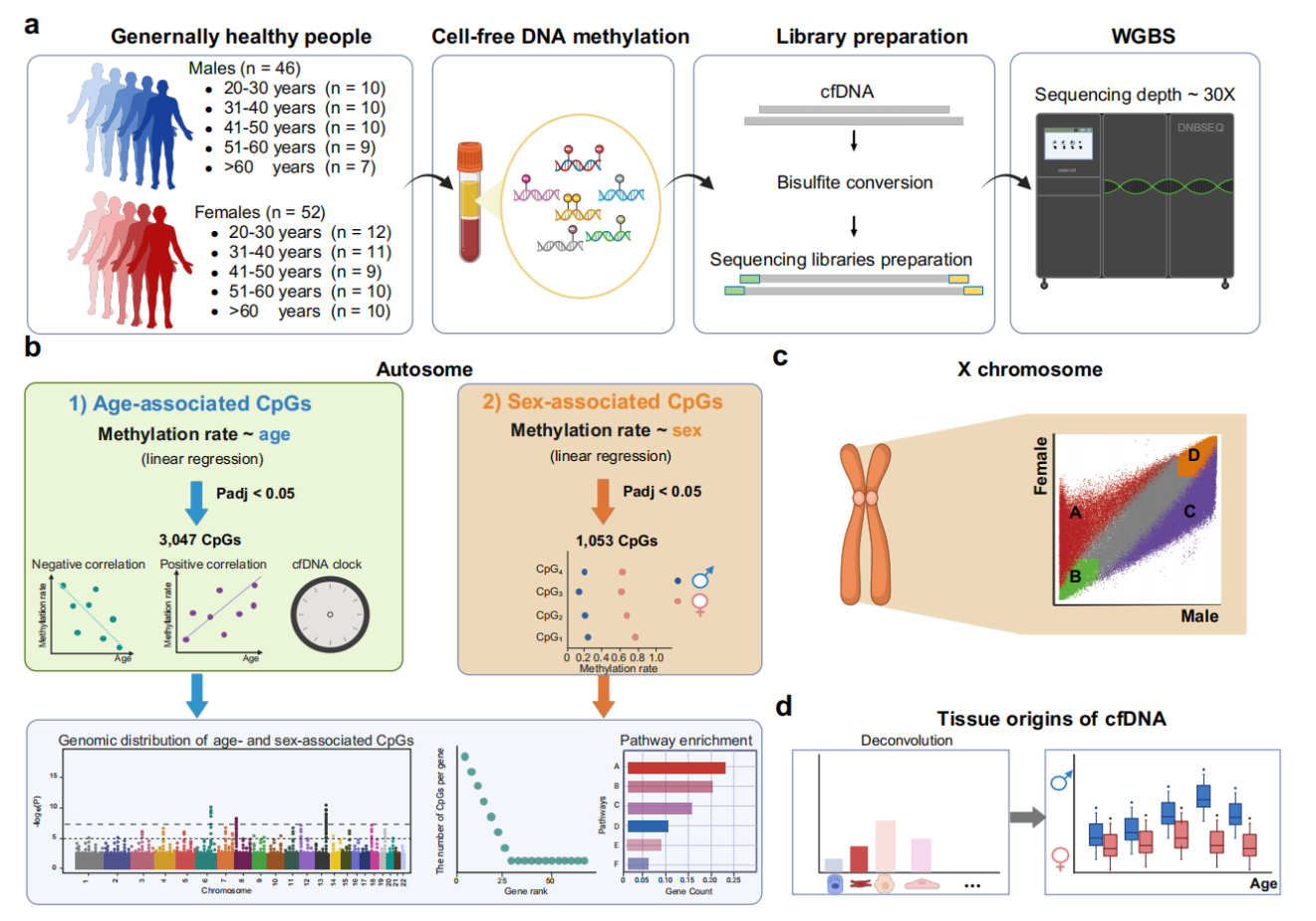
A research team led by BGI-Research reports in Communications Medicine the first comprehensive whole-genome analysis of DNA methylation patterns in plasma cell-free DNA (cfDNA) from 98 generally healthy adults aged 22-77.
DNA methylation, chemical modifications at CpG dinucleotide sites that regulate gene expression, has been linked to aging and sex-biased disease risks. However, existing methylation arrays cover only ~3% of the genome's ~28 million CpG sites, missing most age- and sex-related signals. Using whole-genome bisulfite sequencing (WGBS), this study profiles over 24 million CpG sites, establishing a high-coverage methylation reference and identifying 3,047 age-associated and 1,053 sex-associated CpGs on autosomes, many newly discovered at the DNA methylation level. These findings provide a foundation for distinguishing normal demographic variation from clinically relevant methylation changes in future cfDNA biomarker development.
 Communications Medicine journal website featuring this research on whole-genome cfDNA methylation profiling for age and sex analysis, supporting high-precision epigenetic age clocks with potential for population health monitoring.
Communications Medicine journal website featuring this research on whole-genome cfDNA methylation profiling for age and sex analysis, supporting high-precision epigenetic age clocks with potential for population health monitoring.
 Whole-genome cfDNA methylation profiling reveals age and sex signatures across millions of CpGs, establishing baseline references for future biomarker development and enabling non-invasive health monitoring in general populations.
Whole-genome cfDNA methylation profiling reveals age and sex signatures across millions of CpGs, establishing baseline references for future biomarker development and enabling non-invasive health monitoring in general populations.
Plasma cfDNA offers noninvasive access to epigenetic information from cells throughout the body, but its low abundance and fragmentation present technical challenges for comprehensive methylation analysis. The researchers optimized a single-stranded library preparation workflow for cfDNA, and incorporated MGI DNBSEQ platform-specific adapters, to achieve >30× sequencing depth. After quality control, the dataset covers 23.5 million autosomal CpG sites and 997,000 CpG sites on the X chromosome, representing ~80% of genomic CpGs and enabling genome-wide association analyses at unprecedented scale compared to traditional methods.
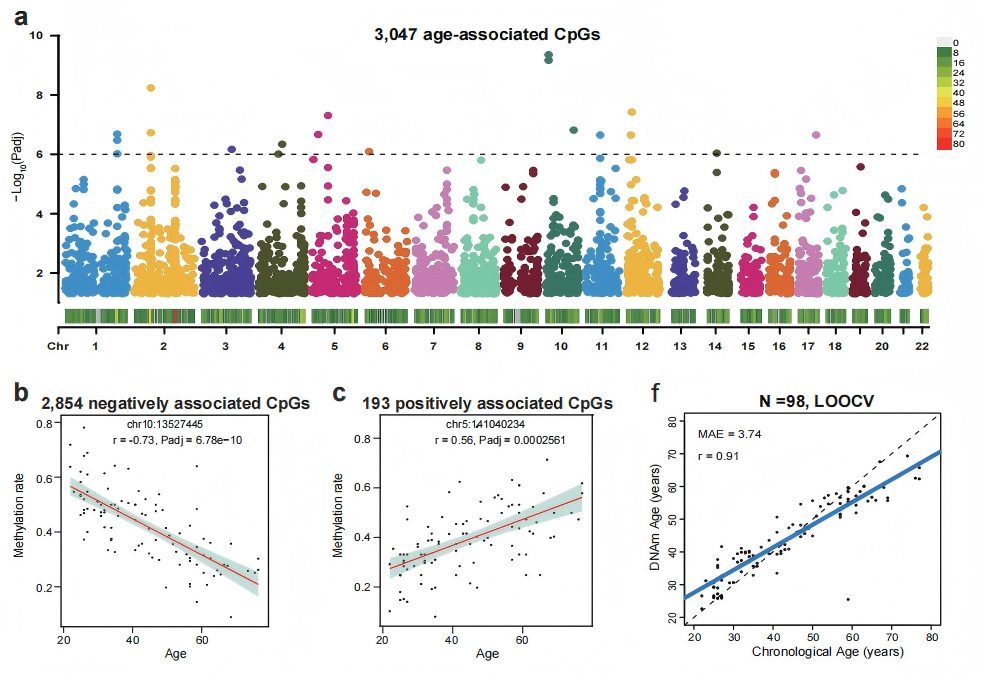 Genome-wide identification of 3,047 age-associated CpGs mapped to 1,587 genes, distilled into a 125-CpG epigenetic clock with 0.91 correlation to chronological age, advancing understanding of aging biology and disease risk assessment.
Genome-wide identification of 3,047 age-associated CpGs mapped to 1,587 genes, distilled into a 125-CpG epigenetic clock with 0.91 correlation to chronological age, advancing understanding of aging biology and disease risk assessment.
The genome-wide analysis identified 3,047 age-associated CpGs (mapping to 1,587 genes) and 1,053 sex-associated CpGs (mapping to 324 genes) on autosomes, with most sites novel discoveries beyond traditional 450K/850K methylation arrays. Age-associated genes include IFT80 (bone health) and RILPL1 (muscle disorders), suggesting new epigenetic mechanisms in age-related diseases. Using elastic net regression, researchers developed a 125-CpG epigenetic clock achieving 0.91 Pearson correlation with chronological age (median absolute error: 3.74 years), validated against external cfDNA WGBS datasets. This demonstrates cfDNA methylation's potential as accessible biomarkers for population-level health monitoring, enabling earlier detection of aging-related risks beyond chronological age alone.
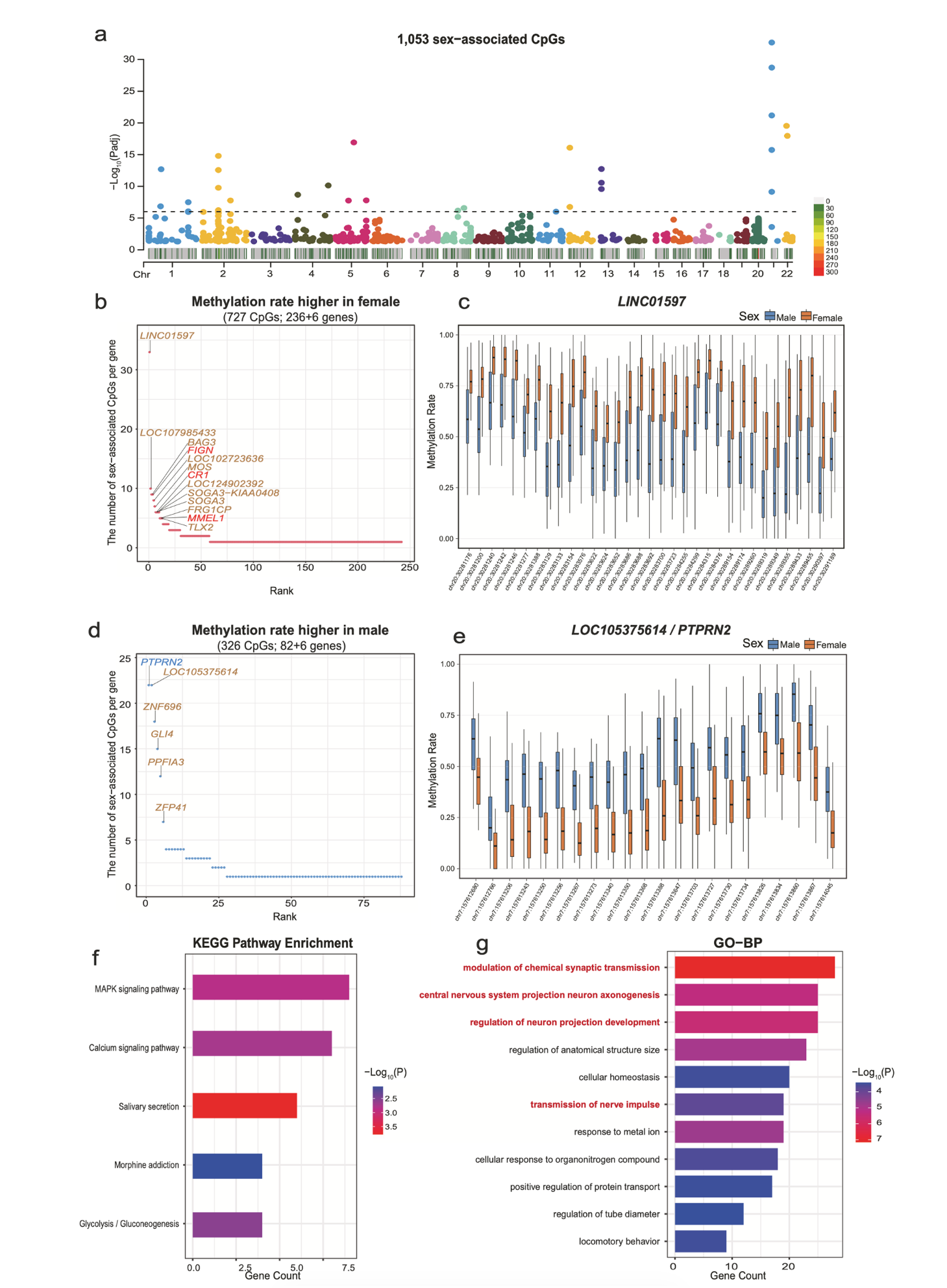 Discovery of 1,053 sex-associated CpGs linked to 324 genes involved in pathways including neural functions and metabolism, providing insights into sex-biased diseases and supporting personalized biomarker development.
Discovery of 1,053 sex-associated CpGs linked to 324 genes involved in pathways including neural functions and metabolism, providing insights into sex-biased diseases and supporting personalized biomarker development.
The study further characterized sex-associated methylation on the X chromosome, revealing patterns consistent with X-chromosome inactivation (XCI) status, including genes that escape inactivation and may contribute to sex-biased autoimmune diseases. Tissue-of-origin deconvolution showed cfDNA is predominantly blood-derived, with hepatocyte contributions associated with liver enzyme markers and age/sex differences. Individual cases with abnormal cell-type proportions (e.g., elevated monocytes in a gout patient, increased granulocytes in those with inflammatory conditions) demonstrate cfDNA's potential for detecting subclinical health issues.
Based on the foundation of the research and extensive data from a large-scale population, a cutting-edge cfDNA epigenetic aging clock model and related technologies have been developed into practical products for biological age assessment. These include a cfDNA methylation-based biological age evaluation using whole-genome sequencing and a targeted methylation testing-based solution. With just a 5mL peripheral venous blood sample, users can receive a comprehensive assessment report covering overall biological age, organ damage risk, disease risk, and health hazards.
Leveraging proprietary high-depth whole-genome methylation sequencing technology, the product fully utilize the unique advantages of cfDNA to simultaneously capture physiological information from multiple organs. The evaluation model provides a scientific basis for precise health management at the genetic level.
The "one-tube multi-omics" technology system enables the in-depth exploration of a single 5-10mL peripheral venous blood sample. Through efficient sample processing, it unlocks multi-dimensional health data, including epigenetic information, transcriptomics, metabolic levels, and protein expression. This truly meets the demand for multiple tests with just one blood sample.
Ethical approval has been obtained for this study. Raw sequencing data and methylation profiles are publicly available for research community use.
This research is available at: https://doi.org/10.1038/s43856-025-01220-y



